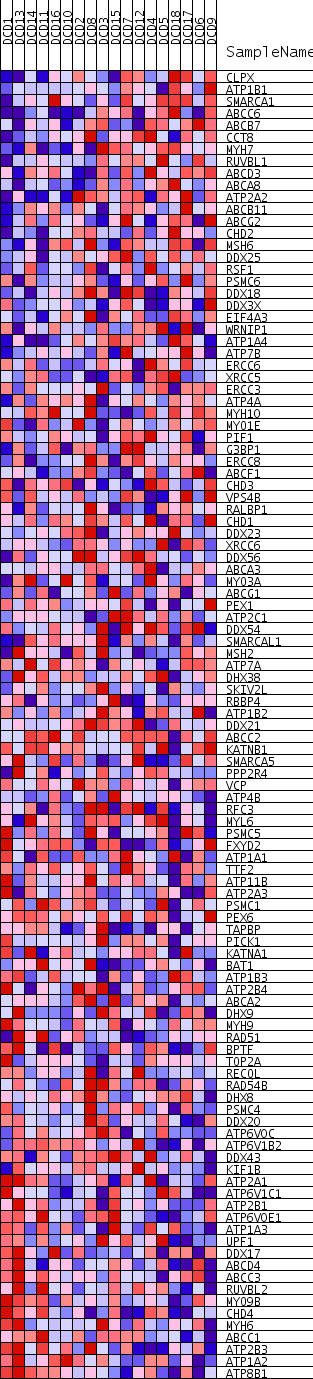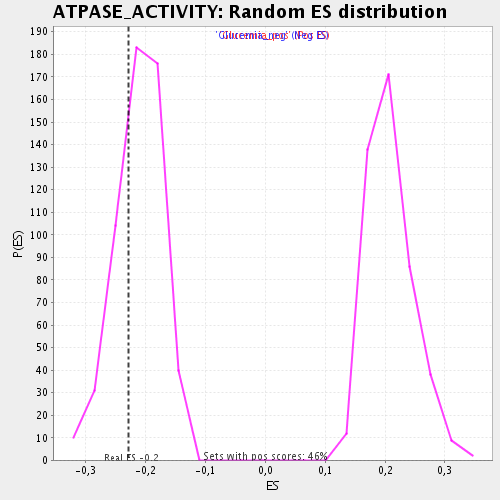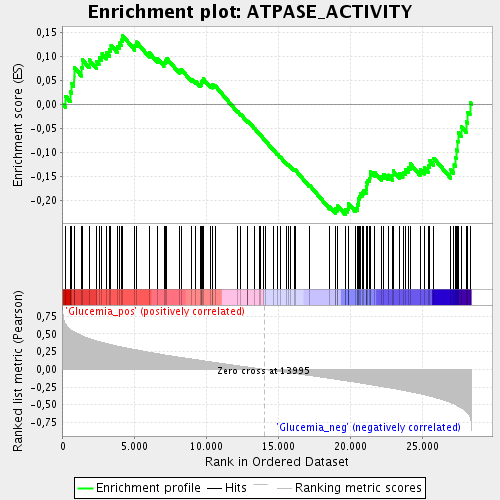
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | DCD_collapsed_to_symbols.DCD.cls#Glucemia |
| Phenotype | DCD.cls#Glucemia |
| Upregulated in class | Glucemia_neg |
| GeneSet | ATPASE_ACTIVITY |
| Enrichment Score (ES) | -0.22868168 |
| Normalized Enrichment Score (NES) | -1.0824946 |
| Nominal p-value | 0.29227942 |
| FDR q-value | 0.8002801 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CLPX | 186 | 0.635 | 0.0173 | No | ||
| 2 | ATP1B1 | 523 | 0.558 | 0.0264 | No | ||
| 3 | SMARCA1 | 624 | 0.544 | 0.0432 | No | ||
| 4 | ABCC6 | 768 | 0.528 | 0.0580 | No | ||
| 5 | ABCB7 | 769 | 0.528 | 0.0778 | No | ||
| 6 | CCT8 | 1300 | 0.476 | 0.0770 | No | ||
| 7 | MYH7 | 1338 | 0.473 | 0.0934 | No | ||
| 8 | RUVBL1 | 1821 | 0.436 | 0.0928 | No | ||
| 9 | ABCD3 | 2350 | 0.401 | 0.0892 | No | ||
| 10 | ABCA8 | 2512 | 0.391 | 0.0982 | No | ||
| 11 | ATP2A2 | 2714 | 0.380 | 0.1053 | No | ||
| 12 | ABCB11 | 3020 | 0.365 | 0.1083 | No | ||
| 13 | ABCG2 | 3239 | 0.355 | 0.1139 | No | ||
| 14 | CHD2 | 3340 | 0.350 | 0.1235 | No | ||
| 15 | MSH6 | 3772 | 0.329 | 0.1206 | No | ||
| 16 | DDX25 | 3910 | 0.323 | 0.1279 | No | ||
| 17 | RSF1 | 4070 | 0.317 | 0.1342 | No | ||
| 18 | PSMC6 | 4164 | 0.313 | 0.1427 | No | ||
| 19 | DDX18 | 5008 | 0.279 | 0.1234 | No | ||
| 20 | DDX3X | 5113 | 0.276 | 0.1300 | No | ||
| 21 | EIF4A3 | 6004 | 0.241 | 0.1077 | No | ||
| 22 | WRNIP1 | 6570 | 0.221 | 0.0960 | No | ||
| 23 | ATP1A4 | 7032 | 0.205 | 0.0874 | No | ||
| 24 | ATP7B | 7111 | 0.202 | 0.0922 | No | ||
| 25 | ERCC6 | 7233 | 0.198 | 0.0954 | No | ||
| 26 | XRCC5 | 8086 | 0.172 | 0.0718 | No | ||
| 27 | ERCC3 | 8251 | 0.167 | 0.0722 | No | ||
| 28 | ATP4A | 8964 | 0.145 | 0.0525 | No | ||
| 29 | MYH10 | 9239 | 0.137 | 0.0480 | No | ||
| 30 | MYO1E | 9556 | 0.127 | 0.0416 | No | ||
| 31 | PIF1 | 9632 | 0.125 | 0.0436 | No | ||
| 32 | G3BP1 | 9646 | 0.124 | 0.0478 | No | ||
| 33 | ERCC8 | 9701 | 0.123 | 0.0505 | No | ||
| 34 | ABCF1 | 9747 | 0.122 | 0.0535 | No | ||
| 35 | CHD3 | 10283 | 0.105 | 0.0385 | No | ||
| 36 | VPS4B | 10404 | 0.101 | 0.0381 | No | ||
| 37 | RALBP1 | 10419 | 0.101 | 0.0414 | No | ||
| 38 | CHD1 | 10599 | 0.096 | 0.0386 | No | ||
| 39 | DDX23 | 12177 | 0.050 | -0.0152 | No | ||
| 40 | XRCC6 | 12377 | 0.045 | -0.0205 | No | ||
| 41 | DDX56 | 12839 | 0.033 | -0.0355 | No | ||
| 42 | ABCA3 | 12870 | 0.032 | -0.0354 | No | ||
| 43 | MYO3A | 12871 | 0.032 | -0.0342 | No | ||
| 44 | ABCG1 | 13322 | 0.019 | -0.0494 | No | ||
| 45 | PEX1 | 13656 | 0.009 | -0.0608 | No | ||
| 46 | ATP2C1 | 13724 | 0.008 | -0.0629 | No | ||
| 47 | DDX54 | 13954 | 0.001 | -0.0709 | No | ||
| 48 | SMARCAL1 | 14073 | -0.002 | -0.0750 | No | ||
| 49 | MSH2 | 14657 | -0.019 | -0.0949 | No | ||
| 50 | ATP7A | 14910 | -0.026 | -0.1028 | No | ||
| 51 | DHX38 | 15120 | -0.032 | -0.1090 | No | ||
| 52 | SKIV2L | 15527 | -0.043 | -0.1217 | No | ||
| 53 | RBBP4 | 15669 | -0.047 | -0.1249 | No | ||
| 54 | ATP1B2 | 15856 | -0.052 | -0.1295 | No | ||
| 55 | DDX21 | 16074 | -0.058 | -0.1350 | No | ||
| 56 | ABCC2 | 16156 | -0.060 | -0.1356 | No | ||
| 57 | KATNB1 | 17148 | -0.087 | -0.1673 | No | ||
| 58 | SMARCA5 | 18564 | -0.127 | -0.2125 | No | ||
| 59 | PPP2R4 | 18977 | -0.141 | -0.2218 | No | ||
| 60 | VCP | 18979 | -0.141 | -0.2166 | No | ||
| 61 | ATP4B | 19117 | -0.145 | -0.2159 | No | ||
| 62 | RFC3 | 19122 | -0.145 | -0.2106 | No | ||
| 63 | MYL6 | 19634 | -0.161 | -0.2227 | Yes | ||
| 64 | PSMC5 | 19681 | -0.162 | -0.2182 | Yes | ||
| 65 | FXYD2 | 19831 | -0.166 | -0.2172 | Yes | ||
| 66 | ATP1A1 | 19844 | -0.167 | -0.2114 | Yes | ||
| 67 | TTF2 | 19864 | -0.168 | -0.2058 | Yes | ||
| 68 | ATP11B | 20341 | -0.181 | -0.2158 | Yes | ||
| 69 | ATP2A3 | 20476 | -0.186 | -0.2135 | Yes | ||
| 70 | PSMC1 | 20512 | -0.187 | -0.2077 | Yes | ||
| 71 | PEX6 | 20554 | -0.189 | -0.2021 | Yes | ||
| 72 | TAPBP | 20576 | -0.189 | -0.1957 | Yes | ||
| 73 | PICK1 | 20645 | -0.192 | -0.1909 | Yes | ||
| 74 | KATNA1 | 20683 | -0.193 | -0.1850 | Yes | ||
| 75 | BAT1 | 20833 | -0.198 | -0.1828 | Yes | ||
| 76 | ATP1B3 | 20927 | -0.201 | -0.1786 | Yes | ||
| 77 | ATP2B4 | 21089 | -0.206 | -0.1765 | Yes | ||
| 78 | ABCA2 | 21122 | -0.207 | -0.1699 | Yes | ||
| 79 | DHX9 | 21144 | -0.208 | -0.1628 | Yes | ||
| 80 | MYH9 | 21216 | -0.210 | -0.1574 | Yes | ||
| 81 | RAD51 | 21318 | -0.213 | -0.1530 | Yes | ||
| 82 | BPTF | 21362 | -0.215 | -0.1464 | Yes | ||
| 83 | TOP2A | 21373 | -0.215 | -0.1387 | Yes | ||
| 84 | RECQL | 21659 | -0.225 | -0.1403 | Yes | ||
| 85 | RAD54B | 22175 | -0.242 | -0.1494 | Yes | ||
| 86 | DHX8 | 22313 | -0.247 | -0.1449 | Yes | ||
| 87 | PSMC4 | 22630 | -0.258 | -0.1464 | Yes | ||
| 88 | DDX20 | 22956 | -0.270 | -0.1478 | Yes | ||
| 89 | ATP6V0C | 22962 | -0.270 | -0.1378 | Yes | ||
| 90 | ATP6V1B2 | 23405 | -0.287 | -0.1427 | Yes | ||
| 91 | DDX43 | 23658 | -0.297 | -0.1404 | Yes | ||
| 92 | KIF1B | 23828 | -0.302 | -0.1351 | Yes | ||
| 93 | ATP2A1 | 24023 | -0.310 | -0.1303 | Yes | ||
| 94 | ATP6V1C1 | 24150 | -0.316 | -0.1229 | Yes | ||
| 95 | ATP2B1 | 24845 | -0.344 | -0.1345 | Yes | ||
| 96 | ATP6V0E1 | 25129 | -0.359 | -0.1310 | Yes | ||
| 97 | ATP1A3 | 25429 | -0.375 | -0.1275 | Yes | ||
| 98 | UPF1 | 25523 | -0.379 | -0.1166 | Yes | ||
| 99 | DDX17 | 25803 | -0.394 | -0.1116 | Yes | ||
| 100 | ABCD4 | 26988 | -0.471 | -0.1358 | Yes | ||
| 101 | ABCC3 | 27204 | -0.490 | -0.1250 | Yes | ||
| 102 | RUVBL2 | 27294 | -0.498 | -0.1095 | Yes | ||
| 103 | MYO9B | 27363 | -0.506 | -0.0929 | Yes | ||
| 104 | CHD4 | 27473 | -0.518 | -0.0773 | Yes | ||
| 105 | MYH6 | 27492 | -0.520 | -0.0584 | Yes | ||
| 106 | ABCC1 | 27710 | -0.543 | -0.0457 | Yes | ||
| 107 | ATP2B3 | 28069 | -0.598 | -0.0359 | Yes | ||
| 108 | ATP1A2 | 28155 | -0.618 | -0.0157 | Yes | ||
| 109 | ATP8B1 | 28322 | -0.674 | 0.0037 | Yes |