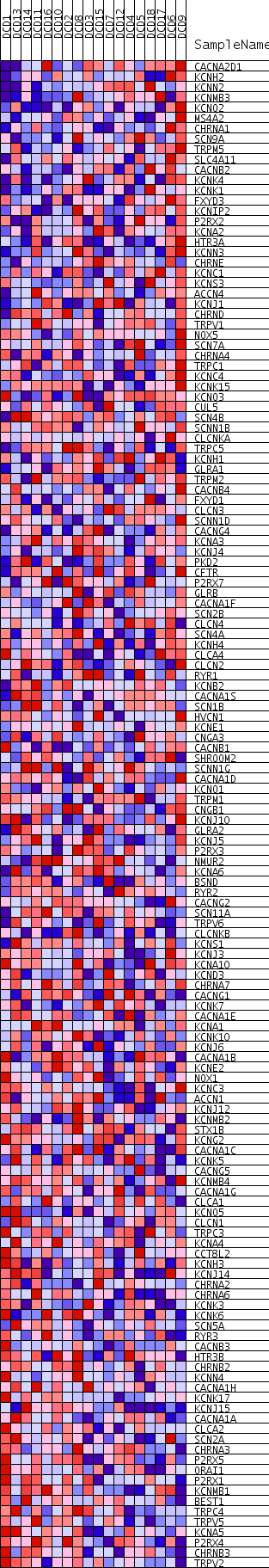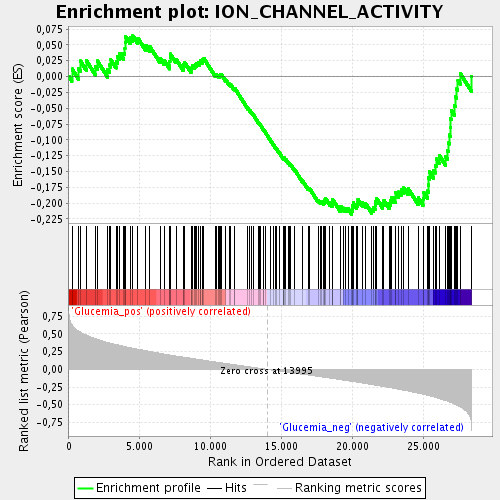
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | DCD_collapsed_to_symbols.DCD.cls#Glucemia |
| Phenotype | DCD.cls#Glucemia |
| Upregulated in class | Glucemia_neg |
| GeneSet | ION_CHANNEL_ACTIVITY |
| Enrichment Score (ES) | -0.2180534 |
| Normalized Enrichment Score (NES) | -1.0780691 |
| Nominal p-value | 0.2870036 |
| FDR q-value | 0.8022616 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CACNA2D1 | 216 | 0.627 | 0.0121 | No | ||
| 2 | KCNH2 | 692 | 0.536 | 0.0123 | No | ||
| 3 | KCNN2 | 806 | 0.525 | 0.0248 | No | ||
| 4 | KCNMB3 | 1223 | 0.483 | 0.0253 | No | ||
| 5 | KCNQ2 | 1872 | 0.432 | 0.0161 | No | ||
| 6 | MS4A2 | 2001 | 0.424 | 0.0249 | No | ||
| 7 | CHRNA1 | 2737 | 0.379 | 0.0109 | No | ||
| 8 | SCN9A | 2856 | 0.374 | 0.0185 | No | ||
| 9 | TRPM5 | 2956 | 0.369 | 0.0266 | No | ||
| 10 | SLC4A11 | 3327 | 0.350 | 0.0246 | No | ||
| 11 | CACNB2 | 3443 | 0.345 | 0.0314 | No | ||
| 12 | KCNK4 | 3581 | 0.339 | 0.0372 | No | ||
| 13 | KCNK1 | 3875 | 0.325 | 0.0371 | No | ||
| 14 | FXYD3 | 3950 | 0.321 | 0.0446 | No | ||
| 15 | KCNIP2 | 3957 | 0.321 | 0.0546 | No | ||
| 16 | P2RX2 | 3984 | 0.320 | 0.0637 | No | ||
| 17 | KCNA2 | 4323 | 0.306 | 0.0614 | No | ||
| 18 | HTR3A | 4508 | 0.299 | 0.0644 | No | ||
| 19 | KCNN3 | 4864 | 0.285 | 0.0608 | No | ||
| 20 | CHRNE | 5417 | 0.264 | 0.0496 | No | ||
| 21 | KCNC1 | 5708 | 0.252 | 0.0473 | No | ||
| 22 | KCNS3 | 6433 | 0.226 | 0.0288 | No | ||
| 23 | ACCN4 | 6729 | 0.216 | 0.0252 | No | ||
| 24 | KCNJ1 | 7105 | 0.202 | 0.0183 | No | ||
| 25 | CHRND | 7110 | 0.202 | 0.0246 | No | ||
| 26 | TRPV1 | 7132 | 0.201 | 0.0302 | No | ||
| 27 | NOX5 | 7151 | 0.201 | 0.0359 | No | ||
| 28 | SCN7A | 7556 | 0.188 | 0.0275 | No | ||
| 29 | CHRNA4 | 8058 | 0.173 | 0.0152 | No | ||
| 30 | TRPC1 | 8092 | 0.172 | 0.0195 | No | ||
| 31 | KCNC4 | 8179 | 0.169 | 0.0218 | No | ||
| 32 | KCNK15 | 8617 | 0.156 | 0.0112 | No | ||
| 33 | KCNQ3 | 8679 | 0.154 | 0.0139 | No | ||
| 34 | CUL5 | 8709 | 0.153 | 0.0177 | No | ||
| 35 | SCN4B | 8841 | 0.149 | 0.0178 | No | ||
| 36 | SCNN1B | 8920 | 0.146 | 0.0197 | No | ||
| 37 | CLCNKA | 9016 | 0.144 | 0.0208 | No | ||
| 38 | TRPC5 | 9109 | 0.140 | 0.0220 | No | ||
| 39 | KCNH1 | 9246 | 0.137 | 0.0215 | No | ||
| 40 | GLRA1 | 9269 | 0.136 | 0.0250 | No | ||
| 41 | TRPM2 | 9396 | 0.132 | 0.0247 | No | ||
| 42 | CACNB4 | 9436 | 0.130 | 0.0275 | No | ||
| 43 | FXYD1 | 9523 | 0.128 | 0.0285 | No | ||
| 44 | CLCN3 | 10331 | 0.103 | 0.0032 | No | ||
| 45 | SCNN1D | 10422 | 0.101 | 0.0032 | No | ||
| 46 | CACNG4 | 10548 | 0.097 | 0.0018 | No | ||
| 47 | KCNA3 | 10652 | 0.094 | 0.0011 | No | ||
| 48 | KCNJ4 | 10679 | 0.093 | 0.0031 | No | ||
| 49 | PKD2 | 10741 | 0.091 | 0.0039 | No | ||
| 50 | CFTR | 11071 | 0.082 | -0.0052 | No | ||
| 51 | P2RX7 | 11319 | 0.075 | -0.0115 | No | ||
| 52 | GLRB | 11418 | 0.072 | -0.0127 | No | ||
| 53 | CACNA1F | 11661 | 0.065 | -0.0192 | No | ||
| 54 | SCN2B | 11700 | 0.064 | -0.0185 | No | ||
| 55 | CLCN4 | 12630 | 0.039 | -0.0502 | No | ||
| 56 | SCN4A | 12764 | 0.035 | -0.0538 | No | ||
| 57 | KCNH4 | 12868 | 0.032 | -0.0564 | No | ||
| 58 | CLCA4 | 13005 | 0.028 | -0.0603 | No | ||
| 59 | CLCN2 | 13402 | 0.017 | -0.0738 | No | ||
| 60 | RYR1 | 13461 | 0.015 | -0.0754 | No | ||
| 61 | KCNB2 | 13507 | 0.014 | -0.0765 | No | ||
| 62 | CACNA1S | 13722 | 0.008 | -0.0839 | No | ||
| 63 | SCN1B | 13727 | 0.007 | -0.0838 | No | ||
| 64 | HVCN1 | 13836 | 0.004 | -0.0875 | No | ||
| 65 | KCNE1 | 14218 | -0.006 | -0.1007 | No | ||
| 66 | CNGA3 | 14446 | -0.012 | -0.1084 | No | ||
| 67 | CACNB1 | 14558 | -0.016 | -0.1118 | No | ||
| 68 | SHROOM2 | 14666 | -0.019 | -0.1150 | No | ||
| 69 | SCNN1G | 14827 | -0.024 | -0.1199 | No | ||
| 70 | CACNA1D | 15126 | -0.032 | -0.1294 | No | ||
| 71 | KCNQ1 | 15155 | -0.033 | -0.1294 | No | ||
| 72 | TRPM1 | 15169 | -0.033 | -0.1288 | No | ||
| 73 | CNGB1 | 15189 | -0.034 | -0.1284 | No | ||
| 74 | KCNJ10 | 15305 | -0.037 | -0.1313 | No | ||
| 75 | GLRA2 | 15456 | -0.041 | -0.1353 | No | ||
| 76 | KCNJ5 | 15587 | -0.045 | -0.1385 | No | ||
| 77 | P2RX3 | 15666 | -0.047 | -0.1398 | No | ||
| 78 | NMUR2 | 15934 | -0.054 | -0.1475 | No | ||
| 79 | KCNA6 | 16483 | -0.069 | -0.1647 | No | ||
| 80 | BSND | 16885 | -0.080 | -0.1763 | No | ||
| 81 | RYR2 | 16969 | -0.082 | -0.1767 | No | ||
| 82 | CACNG2 | 17581 | -0.099 | -0.1952 | No | ||
| 83 | SCN11A | 17735 | -0.103 | -0.1973 | No | ||
| 84 | TRPV6 | 17826 | -0.106 | -0.1972 | No | ||
| 85 | CLCNKB | 17957 | -0.110 | -0.1983 | No | ||
| 86 | KCNS1 | 18027 | -0.111 | -0.1972 | No | ||
| 87 | KCNJ3 | 18063 | -0.112 | -0.1949 | No | ||
| 88 | KCNA10 | 18116 | -0.114 | -0.1932 | No | ||
| 89 | KCND3 | 18401 | -0.123 | -0.1993 | No | ||
| 90 | CHRNA7 | 18574 | -0.127 | -0.2014 | No | ||
| 91 | CACNG1 | 18579 | -0.128 | -0.1975 | No | ||
| 92 | KCNK7 | 18598 | -0.128 | -0.1941 | No | ||
| 93 | CACNA1E | 19145 | -0.146 | -0.2088 | No | ||
| 94 | KCNA1 | 19171 | -0.146 | -0.2051 | No | ||
| 95 | KCNK10 | 19382 | -0.153 | -0.2076 | No | ||
| 96 | KCNJ6 | 19536 | -0.158 | -0.2081 | No | ||
| 97 | CACNA1B | 19704 | -0.163 | -0.2089 | No | ||
| 98 | KCNE2 | 19965 | -0.171 | -0.2127 | Yes | ||
| 99 | NOX1 | 19991 | -0.171 | -0.2081 | Yes | ||
| 100 | KCNC3 | 20002 | -0.172 | -0.2031 | Yes | ||
| 101 | ACCN1 | 20046 | -0.173 | -0.1992 | Yes | ||
| 102 | KCNJ12 | 20298 | -0.180 | -0.2024 | Yes | ||
| 103 | KCNMB2 | 20375 | -0.182 | -0.1993 | Yes | ||
| 104 | STX1B | 20391 | -0.183 | -0.1941 | Yes | ||
| 105 | KCNG2 | 20692 | -0.193 | -0.1986 | Yes | ||
| 106 | CACNA1C | 20933 | -0.201 | -0.2007 | Yes | ||
| 107 | KCNK5 | 21380 | -0.216 | -0.2097 | Yes | ||
| 108 | CACNG5 | 21501 | -0.220 | -0.2070 | Yes | ||
| 109 | KCNMB4 | 21602 | -0.224 | -0.2034 | Yes | ||
| 110 | CACNA1G | 21620 | -0.224 | -0.1970 | Yes | ||
| 111 | CLCA1 | 21712 | -0.227 | -0.1930 | Yes | ||
| 112 | KCNQ5 | 22137 | -0.241 | -0.2004 | Yes | ||
| 113 | CLCN1 | 22224 | -0.244 | -0.1958 | Yes | ||
| 114 | TRPC3 | 22596 | -0.257 | -0.2008 | Yes | ||
| 115 | KCNA4 | 22675 | -0.259 | -0.1954 | Yes | ||
| 116 | CCT8L2 | 22770 | -0.263 | -0.1904 | Yes | ||
| 117 | KCNH3 | 23032 | -0.272 | -0.1910 | Yes | ||
| 118 | KCNJ14 | 23038 | -0.272 | -0.1826 | Yes | ||
| 119 | CHRNA2 | 23280 | -0.281 | -0.1823 | Yes | ||
| 120 | CHRNA6 | 23435 | -0.288 | -0.1786 | Yes | ||
| 121 | KCNK3 | 23622 | -0.295 | -0.1759 | Yes | ||
| 122 | KCNK6 | 23934 | -0.306 | -0.1772 | Yes | ||
| 123 | SCN5A | 24638 | -0.336 | -0.1915 | Yes | ||
| 124 | RYR3 | 24984 | -0.350 | -0.1927 | Yes | ||
| 125 | CACNB3 | 25012 | -0.352 | -0.1825 | Yes | ||
| 126 | HTR3B | 25290 | -0.368 | -0.1807 | Yes | ||
| 127 | CHRNB2 | 25352 | -0.371 | -0.1711 | Yes | ||
| 128 | KCNN4 | 25365 | -0.372 | -0.1598 | Yes | ||
| 129 | CACNA1H | 25436 | -0.375 | -0.1505 | Yes | ||
| 130 | KCNK17 | 25742 | -0.390 | -0.1490 | Yes | ||
| 131 | KCNJ15 | 25842 | -0.396 | -0.1400 | Yes | ||
| 132 | CACNA1A | 25920 | -0.400 | -0.1301 | Yes | ||
| 133 | CLCA2 | 26131 | -0.414 | -0.1244 | Yes | ||
| 134 | SCN2A | 26583 | -0.441 | -0.1265 | Yes | ||
| 135 | CHRNA3 | 26727 | -0.451 | -0.1173 | Yes | ||
| 136 | P2RX5 | 26777 | -0.454 | -0.1047 | Yes | ||
| 137 | ORAI1 | 26832 | -0.459 | -0.0921 | Yes | ||
| 138 | P2RX1 | 26898 | -0.463 | -0.0798 | Yes | ||
| 139 | KCNMB1 | 26920 | -0.464 | -0.0659 | Yes | ||
| 140 | BEST1 | 26989 | -0.471 | -0.0535 | Yes | ||
| 141 | TRPC4 | 27206 | -0.490 | -0.0457 | Yes | ||
| 142 | TRPV5 | 27261 | -0.495 | -0.0320 | Yes | ||
| 143 | KCNA5 | 27358 | -0.505 | -0.0194 | Yes | ||
| 144 | P2RX4 | 27447 | -0.515 | -0.0063 | Yes | ||
| 145 | CHRNB3 | 27605 | -0.532 | 0.0049 | Yes | ||
| 146 | TRPV2 | 28416 | -0.764 | 0.0004 | Yes |