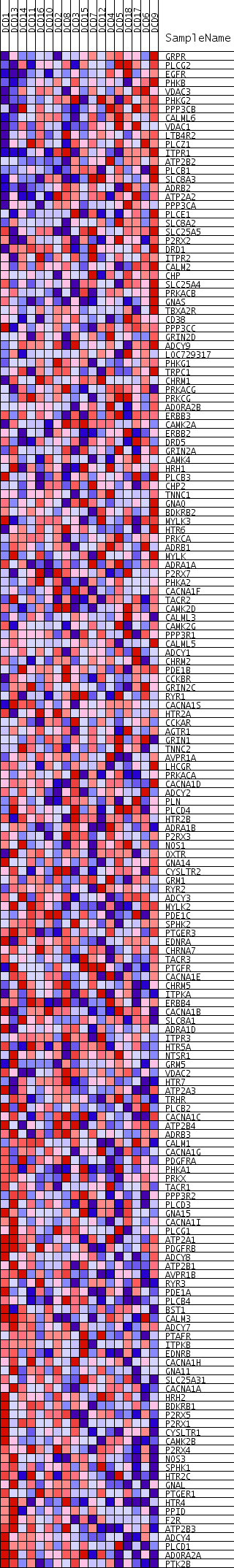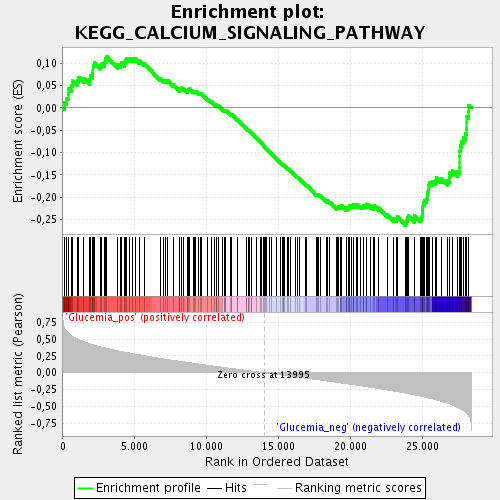
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | DCD_collapsed_to_symbols.DCD.cls#Glucemia |
| Phenotype | DCD.cls#Glucemia |
| Upregulated in class | Glucemia_neg |
| GeneSet | KEGG_CALCIUM_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.26392826 |
| Normalized Enrichment Score (NES) | -1.3487593 |
| Nominal p-value | 0.018450184 |
| FDR q-value | 0.6423613 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GRPR | 100 | 0.670 | 0.0122 | No | ||
| 2 | PLCG2 | 236 | 0.621 | 0.0221 | No | ||
| 3 | EGFR | 359 | 0.593 | 0.0318 | No | ||
| 4 | PHKB | 408 | 0.580 | 0.0437 | No | ||
| 5 | VDAC3 | 614 | 0.545 | 0.0493 | No | ||
| 6 | PHKG2 | 656 | 0.540 | 0.0606 | No | ||
| 7 | PPP3CB | 979 | 0.507 | 0.0612 | No | ||
| 8 | CALML6 | 1108 | 0.494 | 0.0683 | No | ||
| 9 | VDAC1 | 1460 | 0.461 | 0.0667 | No | ||
| 10 | LTB4R2 | 1854 | 0.433 | 0.0630 | No | ||
| 11 | PLCZ1 | 1885 | 0.431 | 0.0721 | No | ||
| 12 | ITPR1 | 2045 | 0.421 | 0.0764 | No | ||
| 13 | ATP2B2 | 2088 | 0.418 | 0.0848 | No | ||
| 14 | PLCB1 | 2090 | 0.418 | 0.0946 | No | ||
| 15 | SLC8A3 | 2175 | 0.412 | 0.1013 | No | ||
| 16 | ADRB2 | 2578 | 0.387 | 0.0962 | No | ||
| 17 | ATP2A2 | 2714 | 0.380 | 0.1004 | No | ||
| 18 | PPP3CA | 2918 | 0.371 | 0.1020 | No | ||
| 19 | PLCE1 | 2939 | 0.370 | 0.1100 | No | ||
| 20 | SLC8A2 | 3029 | 0.365 | 0.1154 | No | ||
| 21 | SLC25A5 | 3790 | 0.329 | 0.0963 | No | ||
| 22 | P2RX2 | 3984 | 0.320 | 0.0970 | No | ||
| 23 | DRD1 | 4053 | 0.317 | 0.1021 | No | ||
| 24 | ITPR2 | 4298 | 0.307 | 0.1007 | No | ||
| 25 | CALM2 | 4344 | 0.305 | 0.1063 | No | ||
| 26 | CHP | 4397 | 0.304 | 0.1116 | No | ||
| 27 | SLC25A4 | 4643 | 0.294 | 0.1098 | No | ||
| 28 | PRKACB | 4837 | 0.286 | 0.1097 | No | ||
| 29 | GNAS | 5022 | 0.279 | 0.1098 | No | ||
| 30 | TBXA2R | 5313 | 0.268 | 0.1058 | No | ||
| 31 | CD38 | 5646 | 0.255 | 0.1001 | No | ||
| 32 | PPP3CC | 6759 | 0.215 | 0.0658 | No | ||
| 33 | GRIN2D | 7026 | 0.205 | 0.0612 | No | ||
| 34 | ADCY9 | 7138 | 0.201 | 0.0620 | No | ||
| 35 | LOC729317 | 7272 | 0.197 | 0.0620 | No | ||
| 36 | PHKG1 | 7659 | 0.185 | 0.0527 | No | ||
| 37 | TRPC1 | 8092 | 0.172 | 0.0414 | No | ||
| 38 | CHRM1 | 8121 | 0.171 | 0.0445 | No | ||
| 39 | PRKACG | 8225 | 0.168 | 0.0448 | No | ||
| 40 | PRKCG | 8378 | 0.163 | 0.0432 | No | ||
| 41 | ADORA2B | 8638 | 0.155 | 0.0377 | No | ||
| 42 | ERBB3 | 8675 | 0.154 | 0.0401 | No | ||
| 43 | CAMK2A | 8714 | 0.152 | 0.0423 | No | ||
| 44 | ERBB2 | 8805 | 0.150 | 0.0427 | No | ||
| 45 | DRD5 | 9067 | 0.142 | 0.0368 | No | ||
| 46 | GRIN2A | 9149 | 0.139 | 0.0372 | No | ||
| 47 | CAMK4 | 9232 | 0.137 | 0.0375 | No | ||
| 48 | HRH1 | 9464 | 0.130 | 0.0324 | No | ||
| 49 | PLCB3 | 9549 | 0.127 | 0.0324 | No | ||
| 50 | CHP2 | 9623 | 0.125 | 0.0328 | No | ||
| 51 | TNNC1 | 10083 | 0.111 | 0.0192 | No | ||
| 52 | GNAQ | 10300 | 0.104 | 0.0140 | No | ||
| 53 | BDKRB2 | 10560 | 0.097 | 0.0071 | No | ||
| 54 | MYLK3 | 10653 | 0.094 | 0.0060 | No | ||
| 55 | HTR6 | 10673 | 0.094 | 0.0076 | No | ||
| 56 | PRKCA | 10791 | 0.090 | 0.0055 | No | ||
| 57 | ADRB1 | 11131 | 0.080 | -0.0046 | No | ||
| 58 | MYLK | 11231 | 0.078 | -0.0062 | No | ||
| 59 | ADRA1A | 11270 | 0.076 | -0.0058 | No | ||
| 60 | P2RX7 | 11319 | 0.075 | -0.0057 | No | ||
| 61 | PHKA2 | 11338 | 0.075 | -0.0046 | No | ||
| 62 | CACNA1F | 11661 | 0.065 | -0.0144 | No | ||
| 63 | TACR2 | 11736 | 0.063 | -0.0156 | No | ||
| 64 | CAMK2D | 11749 | 0.062 | -0.0145 | No | ||
| 65 | CALML3 | 12117 | 0.052 | -0.0263 | No | ||
| 66 | CAMK2G | 12158 | 0.051 | -0.0265 | No | ||
| 67 | PPP3R1 | 12749 | 0.035 | -0.0465 | No | ||
| 68 | CALML5 | 12802 | 0.034 | -0.0476 | No | ||
| 69 | ADCY1 | 12877 | 0.032 | -0.0495 | No | ||
| 70 | CHRM2 | 12933 | 0.030 | -0.0507 | No | ||
| 71 | PDE1B | 12971 | 0.029 | -0.0513 | No | ||
| 72 | CCKBR | 13120 | 0.025 | -0.0560 | No | ||
| 73 | GRIN2C | 13141 | 0.024 | -0.0561 | No | ||
| 74 | RYR1 | 13461 | 0.015 | -0.0670 | No | ||
| 75 | CACNA1S | 13722 | 0.008 | -0.0761 | No | ||
| 76 | HTR2A | 13800 | 0.005 | -0.0787 | No | ||
| 77 | CCKAR | 13964 | 0.001 | -0.0844 | No | ||
| 78 | AGTR1 | 14025 | -0.001 | -0.0865 | No | ||
| 79 | GRIN1 | 14066 | -0.002 | -0.0879 | No | ||
| 80 | TNNC2 | 14152 | -0.005 | -0.0908 | No | ||
| 81 | AVPR1A | 14338 | -0.010 | -0.0971 | No | ||
| 82 | LHCGR | 14489 | -0.014 | -0.1021 | No | ||
| 83 | PRKACA | 14839 | -0.024 | -0.1139 | No | ||
| 84 | CACNA1D | 15126 | -0.032 | -0.1232 | No | ||
| 85 | ADCY2 | 15239 | -0.035 | -0.1264 | No | ||
| 86 | PLN | 15319 | -0.038 | -0.1283 | No | ||
| 87 | PLCD4 | 15324 | -0.038 | -0.1276 | No | ||
| 88 | HTR2B | 15444 | -0.041 | -0.1308 | No | ||
| 89 | ADRA1B | 15588 | -0.045 | -0.1348 | No | ||
| 90 | P2RX3 | 15666 | -0.047 | -0.1364 | No | ||
| 91 | NOS1 | 15855 | -0.052 | -0.1419 | No | ||
| 92 | OXTR | 16160 | -0.061 | -0.1512 | No | ||
| 93 | GNA14 | 16307 | -0.064 | -0.1548 | No | ||
| 94 | CYSLTR2 | 16424 | -0.068 | -0.1573 | No | ||
| 95 | GRM1 | 16891 | -0.080 | -0.1719 | No | ||
| 96 | RYR2 | 16969 | -0.082 | -0.1727 | No | ||
| 97 | ADCY3 | 17645 | -0.101 | -0.1942 | No | ||
| 98 | MYLK2 | 17700 | -0.102 | -0.1937 | No | ||
| 99 | PDE1C | 17760 | -0.104 | -0.1934 | No | ||
| 100 | SPHK2 | 17898 | -0.108 | -0.1957 | No | ||
| 101 | PTGER3 | 18365 | -0.122 | -0.2093 | No | ||
| 102 | EDNRA | 18388 | -0.122 | -0.2072 | No | ||
| 103 | CHRNA7 | 18574 | -0.127 | -0.2107 | No | ||
| 104 | TACR3 | 19032 | -0.142 | -0.2236 | No | ||
| 105 | PTGFR | 19093 | -0.144 | -0.2223 | No | ||
| 106 | CACNA1E | 19145 | -0.146 | -0.2207 | No | ||
| 107 | CHRM5 | 19294 | -0.151 | -0.2223 | No | ||
| 108 | ITPKA | 19321 | -0.152 | -0.2197 | No | ||
| 109 | ERBB4 | 19358 | -0.153 | -0.2174 | No | ||
| 110 | CACNA1B | 19704 | -0.163 | -0.2257 | No | ||
| 111 | SLC8A1 | 19711 | -0.163 | -0.2221 | No | ||
| 112 | ADRA1D | 19888 | -0.168 | -0.2244 | No | ||
| 113 | ITPR3 | 19903 | -0.169 | -0.2209 | No | ||
| 114 | HTR5A | 19926 | -0.170 | -0.2177 | No | ||
| 115 | NTSR1 | 20082 | -0.174 | -0.2190 | No | ||
| 116 | GRM5 | 20211 | -0.177 | -0.2194 | No | ||
| 117 | VDAC2 | 20240 | -0.178 | -0.2162 | No | ||
| 118 | HTR7 | 20455 | -0.185 | -0.2194 | No | ||
| 119 | ATP2A3 | 20476 | -0.186 | -0.2157 | No | ||
| 120 | TRHR | 20733 | -0.195 | -0.2202 | No | ||
| 121 | PLCB2 | 20894 | -0.200 | -0.2212 | No | ||
| 122 | CACNA1C | 20933 | -0.201 | -0.2178 | No | ||
| 123 | ATP2B4 | 21089 | -0.206 | -0.2184 | No | ||
| 124 | ADRB3 | 21129 | -0.207 | -0.2149 | No | ||
| 125 | CALM1 | 21398 | -0.216 | -0.2193 | No | ||
| 126 | CACNA1G | 21620 | -0.224 | -0.2218 | No | ||
| 127 | PDGFRA | 21687 | -0.226 | -0.2188 | No | ||
| 128 | PHKA1 | 21984 | -0.236 | -0.2237 | No | ||
| 129 | PRKX | 22594 | -0.257 | -0.2392 | No | ||
| 130 | TACR1 | 23002 | -0.271 | -0.2472 | No | ||
| 131 | PPP3R2 | 23219 | -0.279 | -0.2483 | No | ||
| 132 | PLCD3 | 23242 | -0.280 | -0.2425 | No | ||
| 133 | GNA15 | 23849 | -0.303 | -0.2568 | Yes | ||
| 134 | CACNA1I | 23900 | -0.305 | -0.2514 | Yes | ||
| 135 | PLCG1 | 23950 | -0.307 | -0.2459 | Yes | ||
| 136 | ATP2A1 | 24023 | -0.310 | -0.2411 | Yes | ||
| 137 | PDGFRB | 24440 | -0.328 | -0.2481 | Yes | ||
| 138 | ADCY8 | 24467 | -0.329 | -0.2413 | Yes | ||
| 139 | ATP2B1 | 24845 | -0.344 | -0.2465 | Yes | ||
| 140 | AVPR1B | 24975 | -0.350 | -0.2428 | Yes | ||
| 141 | RYR3 | 24984 | -0.350 | -0.2349 | Yes | ||
| 142 | PDE1A | 25010 | -0.352 | -0.2274 | Yes | ||
| 143 | PLCB4 | 25044 | -0.354 | -0.2203 | Yes | ||
| 144 | BST1 | 25049 | -0.354 | -0.2121 | Yes | ||
| 145 | CALM3 | 25151 | -0.359 | -0.2072 | Yes | ||
| 146 | ADCY7 | 25294 | -0.368 | -0.2035 | Yes | ||
| 147 | PTAFR | 25341 | -0.370 | -0.1964 | Yes | ||
| 148 | ITPKB | 25357 | -0.371 | -0.1882 | Yes | ||
| 149 | EDNRB | 25415 | -0.374 | -0.1814 | Yes | ||
| 150 | CACNA1H | 25436 | -0.375 | -0.1732 | Yes | ||
| 151 | GNA11 | 25497 | -0.378 | -0.1665 | Yes | ||
| 152 | SLC25A31 | 25677 | -0.387 | -0.1637 | Yes | ||
| 153 | CACNA1A | 25920 | -0.400 | -0.1628 | Yes | ||
| 154 | HRH2 | 25959 | -0.403 | -0.1547 | Yes | ||
| 155 | BDKRB1 | 26330 | -0.426 | -0.1577 | Yes | ||
| 156 | P2RX5 | 26777 | -0.454 | -0.1628 | Yes | ||
| 157 | P2RX1 | 26898 | -0.463 | -0.1561 | Yes | ||
| 158 | CYSLTR1 | 26906 | -0.464 | -0.1455 | Yes | ||
| 159 | CAMK2B | 27073 | -0.478 | -0.1400 | Yes | ||
| 160 | P2RX4 | 27447 | -0.515 | -0.1411 | Yes | ||
| 161 | NOS3 | 27576 | -0.530 | -0.1332 | Yes | ||
| 162 | SPHK1 | 27592 | -0.531 | -0.1212 | Yes | ||
| 163 | HTR2C | 27594 | -0.531 | -0.1087 | Yes | ||
| 164 | GNAL | 27599 | -0.532 | -0.0963 | Yes | ||
| 165 | PTGER1 | 27628 | -0.535 | -0.0847 | Yes | ||
| 166 | HTR4 | 27719 | -0.544 | -0.0750 | Yes | ||
| 167 | PPID | 27850 | -0.561 | -0.0664 | Yes | ||
| 168 | F2R | 28002 | -0.586 | -0.0579 | Yes | ||
| 169 | ATP2B3 | 28069 | -0.598 | -0.0462 | Yes | ||
| 170 | ADCY4 | 28105 | -0.606 | -0.0332 | Yes | ||
| 171 | PLCD1 | 28107 | -0.606 | -0.0189 | Yes | ||
| 172 | ADORA2A | 28218 | -0.635 | -0.0078 | Yes | ||
| 173 | PTK2B | 28245 | -0.644 | 0.0064 | Yes |