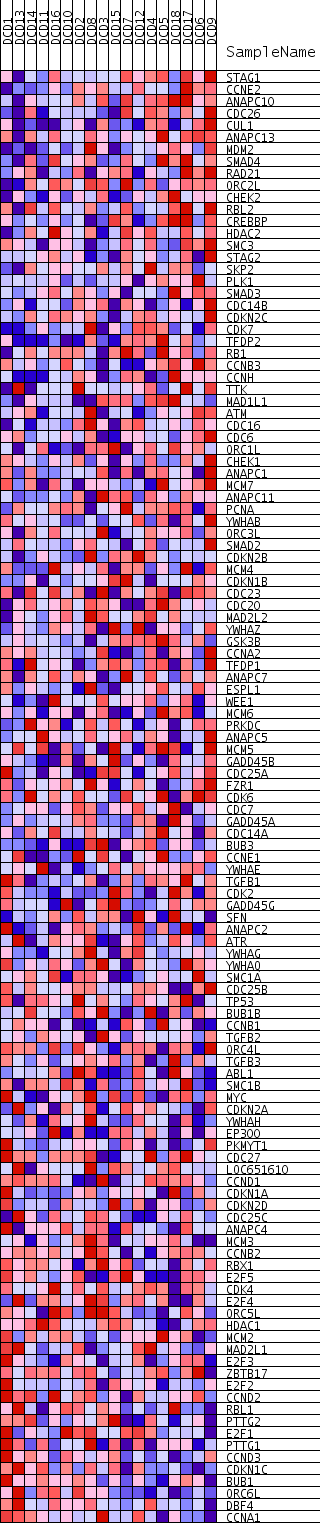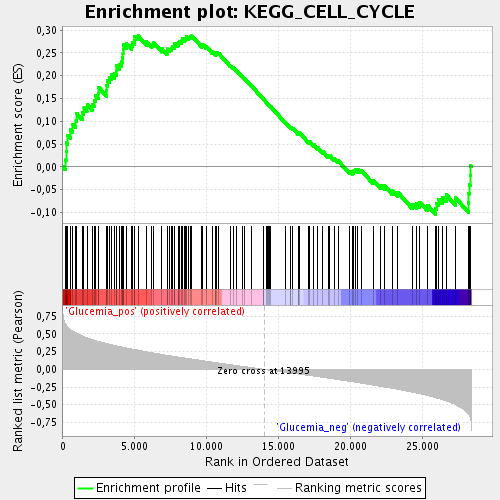
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | DCD_collapsed_to_symbols.DCD.cls#Glucemia |
| Phenotype | DCD.cls#Glucemia |
| Upregulated in class | Glucemia_pos |
| GeneSet | KEGG_CELL_CYCLE |
| Enrichment Score (ES) | 0.2871443 |
| Normalized Enrichment Score (NES) | 1.406495 |
| Nominal p-value | 0.00998004 |
| FDR q-value | 0.19922751 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | STAG1 | 168 | 0.642 | 0.0151 | Yes | ||
| 2 | CCNE2 | 215 | 0.628 | 0.0339 | Yes | ||
| 3 | ANAPC10 | 240 | 0.620 | 0.0534 | Yes | ||
| 4 | CDC26 | 344 | 0.595 | 0.0692 | Yes | ||
| 5 | CUL1 | 521 | 0.558 | 0.0812 | Yes | ||
| 6 | ANAPC13 | 681 | 0.537 | 0.0931 | Yes | ||
| 7 | MDM2 | 906 | 0.515 | 0.1020 | Yes | ||
| 8 | SMAD4 | 969 | 0.507 | 0.1164 | Yes | ||
| 9 | RAD21 | 1347 | 0.472 | 0.1185 | Yes | ||
| 10 | ORC2L | 1463 | 0.461 | 0.1295 | Yes | ||
| 11 | CHEK2 | 1676 | 0.445 | 0.1366 | Yes | ||
| 12 | RBL2 | 2052 | 0.420 | 0.1371 | Yes | ||
| 13 | CREBBP | 2169 | 0.412 | 0.1464 | Yes | ||
| 14 | HDAC2 | 2273 | 0.406 | 0.1561 | Yes | ||
| 15 | SMC3 | 2477 | 0.393 | 0.1617 | Yes | ||
| 16 | STAG2 | 2484 | 0.393 | 0.1744 | Yes | ||
| 17 | SKP2 | 3030 | 0.365 | 0.1670 | Yes | ||
| 18 | PLK1 | 3050 | 0.364 | 0.1782 | Yes | ||
| 19 | SMAD3 | 3094 | 0.362 | 0.1885 | Yes | ||
| 20 | CDC14B | 3218 | 0.356 | 0.1958 | Yes | ||
| 21 | CDKN2C | 3360 | 0.348 | 0.2022 | Yes | ||
| 22 | CDK7 | 3584 | 0.338 | 0.2054 | Yes | ||
| 23 | TFDP2 | 3708 | 0.332 | 0.2119 | Yes | ||
| 24 | RB1 | 3741 | 0.331 | 0.2216 | Yes | ||
| 25 | CCNB3 | 3952 | 0.321 | 0.2247 | Yes | ||
| 26 | CCNH | 4095 | 0.315 | 0.2300 | Yes | ||
| 27 | TTK | 4129 | 0.314 | 0.2391 | Yes | ||
| 28 | MAD1L1 | 4175 | 0.313 | 0.2477 | Yes | ||
| 29 | ATM | 4180 | 0.312 | 0.2578 | Yes | ||
| 30 | CDC16 | 4195 | 0.312 | 0.2675 | Yes | ||
| 31 | CDC6 | 4403 | 0.303 | 0.2701 | Yes | ||
| 32 | ORC1L | 4771 | 0.289 | 0.2666 | Yes | ||
| 33 | CHEK1 | 4848 | 0.285 | 0.2732 | Yes | ||
| 34 | ANAPC1 | 4971 | 0.280 | 0.2780 | Yes | ||
| 35 | MCM7 | 4994 | 0.280 | 0.2864 | Yes | ||
| 36 | ANAPC11 | 5225 | 0.271 | 0.2871 | Yes | ||
| 37 | PCNA | 5819 | 0.248 | 0.2743 | No | ||
| 38 | YWHAB | 6190 | 0.234 | 0.2689 | No | ||
| 39 | ORC3L | 6307 | 0.230 | 0.2723 | No | ||
| 40 | SMAD2 | 6889 | 0.210 | 0.2587 | No | ||
| 41 | CDKN2B | 7241 | 0.198 | 0.2527 | No | ||
| 42 | MCM4 | 7262 | 0.197 | 0.2585 | No | ||
| 43 | CDKN1B | 7443 | 0.192 | 0.2584 | No | ||
| 44 | CDC23 | 7562 | 0.188 | 0.2603 | No | ||
| 45 | CDC20 | 7606 | 0.187 | 0.2649 | No | ||
| 46 | MAD2L2 | 7743 | 0.182 | 0.2661 | No | ||
| 47 | YWHAZ | 7786 | 0.181 | 0.2705 | No | ||
| 48 | GSK3B | 8005 | 0.174 | 0.2685 | No | ||
| 49 | CCNA2 | 8054 | 0.173 | 0.2725 | No | ||
| 50 | TFDP1 | 8124 | 0.171 | 0.2756 | No | ||
| 51 | ANAPC7 | 8268 | 0.167 | 0.2760 | No | ||
| 52 | ESPL1 | 8292 | 0.165 | 0.2806 | No | ||
| 53 | WEE1 | 8476 | 0.160 | 0.2794 | No | ||
| 54 | MCM6 | 8540 | 0.158 | 0.2823 | No | ||
| 55 | PRKDC | 8561 | 0.158 | 0.2867 | No | ||
| 56 | ANAPC5 | 8764 | 0.151 | 0.2845 | No | ||
| 57 | MCM5 | 8857 | 0.149 | 0.2862 | No | ||
| 58 | GADD45B | 8966 | 0.145 | 0.2871 | No | ||
| 59 | CDC25A | 9666 | 0.124 | 0.2664 | No | ||
| 60 | FZR1 | 9718 | 0.122 | 0.2686 | No | ||
| 61 | CDK6 | 9978 | 0.115 | 0.2632 | No | ||
| 62 | CDC7 | 10386 | 0.102 | 0.2522 | No | ||
| 63 | GADD45A | 10617 | 0.095 | 0.2471 | No | ||
| 64 | CDC14A | 10640 | 0.094 | 0.2494 | No | ||
| 65 | BUB3 | 10669 | 0.094 | 0.2515 | No | ||
| 66 | CCNE1 | 10821 | 0.089 | 0.2491 | No | ||
| 67 | YWHAE | 11672 | 0.065 | 0.2212 | No | ||
| 68 | TGFB1 | 11836 | 0.060 | 0.2174 | No | ||
| 69 | CDK2 | 12058 | 0.054 | 0.2114 | No | ||
| 70 | GADD45G | 12493 | 0.042 | 0.1974 | No | ||
| 71 | SFN | 12639 | 0.039 | 0.1935 | No | ||
| 72 | ANAPC2 | 13137 | 0.024 | 0.1768 | No | ||
| 73 | ATR | 13923 | 0.002 | 0.1491 | No | ||
| 74 | YWHAG | 13973 | 0.001 | 0.1474 | No | ||
| 75 | YWHAQ | 14180 | -0.005 | 0.1403 | No | ||
| 76 | SMC1A | 14219 | -0.007 | 0.1391 | No | ||
| 77 | CDC25B | 14293 | -0.009 | 0.1368 | No | ||
| 78 | TP53 | 14396 | -0.011 | 0.1336 | No | ||
| 79 | BUB1B | 14467 | -0.013 | 0.1316 | No | ||
| 80 | CCNB1 | 15516 | -0.043 | 0.0959 | No | ||
| 81 | TGFB2 | 15828 | -0.051 | 0.0866 | No | ||
| 82 | ORC4L | 15859 | -0.052 | 0.0873 | No | ||
| 83 | TGFB3 | 15996 | -0.056 | 0.0843 | No | ||
| 84 | ABL1 | 16389 | -0.067 | 0.0726 | No | ||
| 85 | SMC1B | 16403 | -0.067 | 0.0744 | No | ||
| 86 | MYC | 16431 | -0.068 | 0.0756 | No | ||
| 87 | CDKN2A | 17105 | -0.086 | 0.0547 | No | ||
| 88 | YWHAH | 17155 | -0.087 | 0.0558 | No | ||
| 89 | EP300 | 17453 | -0.096 | 0.0484 | No | ||
| 90 | PKMYT1 | 17736 | -0.103 | 0.0419 | No | ||
| 91 | CDC27 | 18074 | -0.113 | 0.0336 | No | ||
| 92 | LOC651610 | 18465 | -0.124 | 0.0239 | No | ||
| 93 | CCND1 | 18562 | -0.127 | 0.0247 | No | ||
| 94 | CDKN1A | 18876 | -0.137 | 0.0181 | No | ||
| 95 | CDKN2D | 19146 | -0.146 | 0.0134 | No | ||
| 96 | CDC25C | 19946 | -0.170 | -0.0093 | No | ||
| 97 | ANAPC4 | 20141 | -0.176 | -0.0104 | No | ||
| 98 | MCM3 | 20246 | -0.178 | -0.0083 | No | ||
| 99 | CCNB2 | 20332 | -0.181 | -0.0054 | No | ||
| 100 | RBX1 | 20523 | -0.188 | -0.0059 | No | ||
| 101 | E2F5 | 20735 | -0.195 | -0.0070 | No | ||
| 102 | CDK4 | 21574 | -0.223 | -0.0294 | No | ||
| 103 | E2F4 | 22102 | -0.240 | -0.0401 | No | ||
| 104 | ORC5L | 22368 | -0.249 | -0.0413 | No | ||
| 105 | HDAC1 | 22926 | -0.268 | -0.0522 | No | ||
| 106 | MCM2 | 23297 | -0.282 | -0.0561 | No | ||
| 107 | MAD2L1 | 24318 | -0.322 | -0.0816 | No | ||
| 108 | E2F3 | 24568 | -0.333 | -0.0795 | No | ||
| 109 | ZBTB17 | 24818 | -0.343 | -0.0771 | No | ||
| 110 | E2F2 | 25370 | -0.372 | -0.0844 | No | ||
| 111 | CCND2 | 25943 | -0.402 | -0.0915 | No | ||
| 112 | RBL1 | 26013 | -0.407 | -0.0806 | No | ||
| 113 | PTTG2 | 26122 | -0.413 | -0.0709 | No | ||
| 114 | E2F1 | 26409 | -0.430 | -0.0670 | No | ||
| 115 | PTTG1 | 26662 | -0.446 | -0.0613 | No | ||
| 116 | CCND3 | 27316 | -0.500 | -0.0681 | No | ||
| 117 | CDKN1C | 28206 | -0.632 | -0.0788 | No | ||
| 118 | BUB1 | 28237 | -0.642 | -0.0589 | No | ||
| 119 | ORC6L | 28257 | -0.648 | -0.0384 | No | ||
| 120 | DBF4 | 28325 | -0.676 | -0.0187 | No | ||
| 121 | CCNA1 | 28335 | -0.680 | 0.0033 | No |