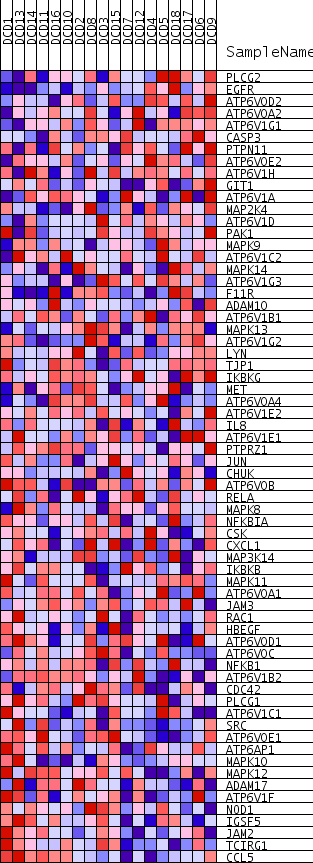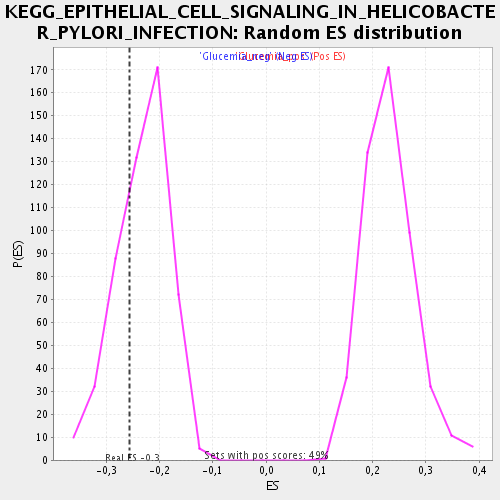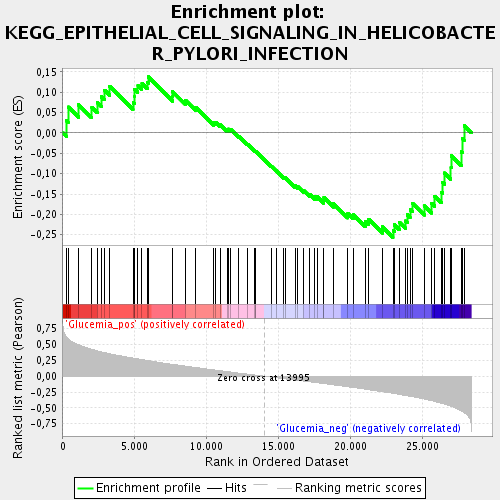
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | DCD_collapsed_to_symbols.DCD.cls#Glucemia |
| Phenotype | DCD.cls#Glucemia |
| Upregulated in class | Glucemia_neg |
| GeneSet | KEGG_EPITHELIAL_CELL_SIGNALING_IN_HELICOBACTER_PYLORI_INFECTION |
| Enrichment Score (ES) | -0.2562394 |
| Normalized Enrichment Score (NES) | -1.1061718 |
| Nominal p-value | 0.28627452 |
| FDR q-value | 0.7898931 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PLCG2 | 236 | 0.621 | 0.0308 | No | ||
| 2 | EGFR | 359 | 0.593 | 0.0637 | No | ||
| 3 | ATP6V0D2 | 1075 | 0.497 | 0.0698 | No | ||
| 4 | ATP6V0A2 | 2015 | 0.423 | 0.0633 | No | ||
| 5 | ATP6V1G1 | 2416 | 0.397 | 0.0742 | No | ||
| 6 | CASP3 | 2677 | 0.382 | 0.0891 | No | ||
| 7 | PTPN11 | 2909 | 0.371 | 0.1043 | No | ||
| 8 | ATP6V0E2 | 3260 | 0.354 | 0.1142 | No | ||
| 9 | ATP6V1H | 4875 | 0.284 | 0.0752 | No | ||
| 10 | GIT1 | 4960 | 0.281 | 0.0899 | No | ||
| 11 | ATP6V1A | 4967 | 0.281 | 0.1074 | No | ||
| 12 | MAP2K4 | 5219 | 0.271 | 0.1156 | No | ||
| 13 | ATP6V1D | 5497 | 0.261 | 0.1222 | No | ||
| 14 | PAK1 | 5877 | 0.246 | 0.1243 | No | ||
| 15 | MAPK9 | 5933 | 0.244 | 0.1377 | No | ||
| 16 | ATP6V1C2 | 7626 | 0.186 | 0.0898 | No | ||
| 17 | MAPK14 | 7635 | 0.186 | 0.1012 | No | ||
| 18 | ATP6V1G3 | 8537 | 0.158 | 0.0794 | No | ||
| 19 | F11R | 9248 | 0.137 | 0.0630 | No | ||
| 20 | ADAM10 | 10494 | 0.098 | 0.0253 | No | ||
| 21 | ATP6V1B1 | 10642 | 0.094 | 0.0260 | No | ||
| 22 | MAPK13 | 10959 | 0.085 | 0.0202 | No | ||
| 23 | ATP6V1G2 | 11462 | 0.071 | 0.0070 | No | ||
| 24 | LYN | 11511 | 0.070 | 0.0097 | No | ||
| 25 | TJP1 | 11663 | 0.065 | 0.0085 | No | ||
| 26 | IKBKG | 12234 | 0.049 | -0.0085 | No | ||
| 27 | MET | 12825 | 0.033 | -0.0272 | No | ||
| 28 | ATP6V0A4 | 13343 | 0.019 | -0.0443 | No | ||
| 29 | ATP6V1E2 | 13398 | 0.017 | -0.0452 | No | ||
| 30 | IL8 | 13429 | 0.016 | -0.0452 | No | ||
| 31 | ATP6V1E1 | 14509 | -0.014 | -0.0823 | No | ||
| 32 | PTPRZ1 | 14851 | -0.025 | -0.0928 | No | ||
| 33 | JUN | 15375 | -0.039 | -0.1088 | No | ||
| 34 | CHUK | 15476 | -0.042 | -0.1097 | No | ||
| 35 | ATP6V0B | 16150 | -0.060 | -0.1297 | No | ||
| 36 | RELA | 16345 | -0.066 | -0.1324 | No | ||
| 37 | MAPK8 | 16763 | -0.077 | -0.1423 | No | ||
| 38 | NFKBIA | 17179 | -0.088 | -0.1513 | No | ||
| 39 | CSK | 17503 | -0.097 | -0.1566 | No | ||
| 40 | CXCL1 | 17677 | -0.102 | -0.1563 | No | ||
| 41 | MAP3K14 | 18133 | -0.114 | -0.1652 | No | ||
| 42 | IKBKB | 18157 | -0.115 | -0.1587 | No | ||
| 43 | MAPK11 | 18822 | -0.135 | -0.1736 | No | ||
| 44 | ATP6V0A1 | 19825 | -0.166 | -0.1985 | No | ||
| 45 | JAM3 | 20190 | -0.177 | -0.2002 | No | ||
| 46 | RAC1 | 21030 | -0.204 | -0.2170 | No | ||
| 47 | HBEGF | 21278 | -0.212 | -0.2123 | No | ||
| 48 | ATP6V0D1 | 22234 | -0.245 | -0.2306 | No | ||
| 49 | ATP6V0C | 22962 | -0.270 | -0.2393 | Yes | ||
| 50 | NFKB1 | 23048 | -0.273 | -0.2251 | Yes | ||
| 51 | ATP6V1B2 | 23405 | -0.287 | -0.2196 | Yes | ||
| 52 | CDC42 | 23850 | -0.303 | -0.2162 | Yes | ||
| 53 | PLCG1 | 23950 | -0.307 | -0.2004 | Yes | ||
| 54 | ATP6V1C1 | 24150 | -0.316 | -0.1875 | Yes | ||
| 55 | SRC | 24313 | -0.322 | -0.1730 | Yes | ||
| 56 | ATP6V0E1 | 25129 | -0.359 | -0.1792 | Yes | ||
| 57 | ATP6AP1 | 25631 | -0.384 | -0.1726 | Yes | ||
| 58 | MAPK10 | 25860 | -0.397 | -0.1557 | Yes | ||
| 59 | MAPK12 | 26321 | -0.425 | -0.1452 | Yes | ||
| 60 | ADAM17 | 26417 | -0.431 | -0.1214 | Yes | ||
| 61 | ATP6V1F | 26545 | -0.439 | -0.0983 | Yes | ||
| 62 | NOD1 | 26995 | -0.471 | -0.0845 | Yes | ||
| 63 | IGSF5 | 27016 | -0.473 | -0.0554 | Yes | ||
| 64 | JAM2 | 27736 | -0.546 | -0.0464 | Yes | ||
| 65 | TCIRG1 | 27782 | -0.553 | -0.0132 | Yes | ||
| 66 | CCL5 | 27908 | -0.571 | 0.0183 | Yes |