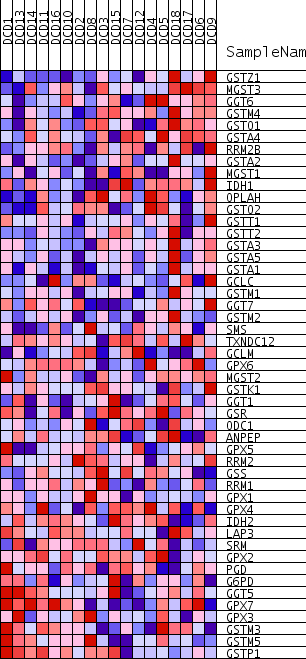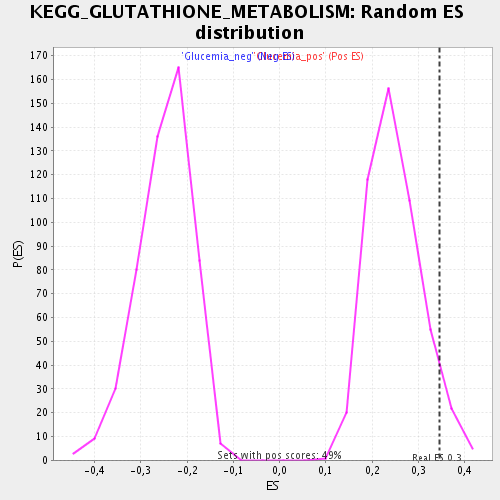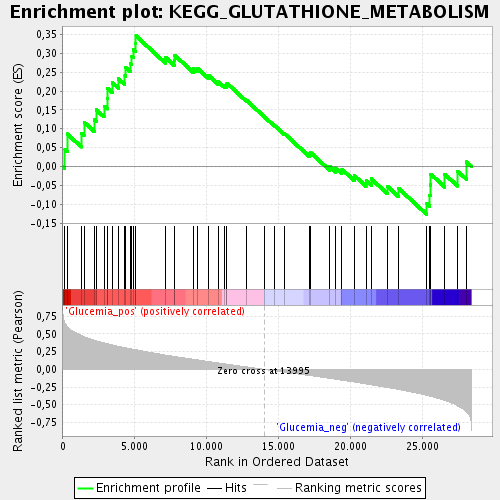
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | DCD_collapsed_to_symbols.DCD.cls#Glucemia |
| Phenotype | DCD.cls#Glucemia |
| Upregulated in class | Glucemia_pos |
| GeneSet | KEGG_GLUTATHIONE_METABOLISM |
| Enrichment Score (ES) | 0.3460525 |
| Normalized Enrichment Score (NES) | 1.3935298 |
| Nominal p-value | 0.059670784 |
| FDR q-value | 0.20919706 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GSTZ1 | 140 | 0.652 | 0.0453 | Yes | ||
| 2 | MGST3 | 302 | 0.605 | 0.0863 | Yes | ||
| 3 | GGT6 | 1286 | 0.477 | 0.0884 | Yes | ||
| 4 | GSTM4 | 1469 | 0.461 | 0.1175 | Yes | ||
| 5 | GSTO1 | 2196 | 0.411 | 0.1236 | Yes | ||
| 6 | GSTA4 | 2323 | 0.403 | 0.1502 | Yes | ||
| 7 | RRM2B | 2863 | 0.373 | 0.1600 | Yes | ||
| 8 | GSTA2 | 3081 | 0.362 | 0.1803 | Yes | ||
| 9 | MGST1 | 3100 | 0.361 | 0.2075 | Yes | ||
| 10 | IDH1 | 3427 | 0.345 | 0.2226 | Yes | ||
| 11 | OPLAH | 3862 | 0.325 | 0.2324 | Yes | ||
| 12 | GSTO2 | 4314 | 0.306 | 0.2401 | Yes | ||
| 13 | GSTT1 | 4360 | 0.305 | 0.2621 | Yes | ||
| 14 | GSTT2 | 4682 | 0.293 | 0.2733 | Yes | ||
| 15 | GSTA3 | 4795 | 0.287 | 0.2915 | Yes | ||
| 16 | GSTA5 | 4903 | 0.283 | 0.3095 | Yes | ||
| 17 | GSTA1 | 5053 | 0.278 | 0.3257 | Yes | ||
| 18 | GCLC | 5081 | 0.276 | 0.3461 | Yes | ||
| 19 | GSTM1 | 7127 | 0.201 | 0.2895 | No | ||
| 20 | GGT7 | 7753 | 0.182 | 0.2815 | No | ||
| 21 | GSTM2 | 7781 | 0.181 | 0.2946 | No | ||
| 22 | SMS | 9092 | 0.141 | 0.2593 | No | ||
| 23 | TXNDC12 | 9344 | 0.134 | 0.2608 | No | ||
| 24 | GCLM | 10134 | 0.110 | 0.2414 | No | ||
| 25 | GPX6 | 10804 | 0.089 | 0.2247 | No | ||
| 26 | MGST2 | 11248 | 0.077 | 0.2150 | No | ||
| 27 | GSTK1 | 11408 | 0.073 | 0.2150 | No | ||
| 28 | GGT1 | 11409 | 0.073 | 0.2207 | No | ||
| 29 | GSR | 12801 | 0.034 | 0.1743 | No | ||
| 30 | ODC1 | 14048 | -0.002 | 0.1305 | No | ||
| 31 | ANPEP | 14751 | -0.022 | 0.1074 | No | ||
| 32 | GPX5 | 15409 | -0.040 | 0.0873 | No | ||
| 33 | RRM2 | 17130 | -0.087 | 0.0334 | No | ||
| 34 | GSS | 17246 | -0.090 | 0.0363 | No | ||
| 35 | RRM1 | 18555 | -0.127 | -0.0000 | No | ||
| 36 | GPX1 | 18973 | -0.140 | -0.0039 | No | ||
| 37 | GPX4 | 19410 | -0.154 | -0.0074 | No | ||
| 38 | IDH2 | 20270 | -0.179 | -0.0238 | No | ||
| 39 | LAP3 | 21087 | -0.206 | -0.0367 | No | ||
| 40 | SRM | 21447 | -0.218 | -0.0325 | No | ||
| 41 | GPX2 | 22570 | -0.256 | -0.0523 | No | ||
| 42 | PGD | 23368 | -0.285 | -0.0584 | No | ||
| 43 | G6PD | 25323 | -0.369 | -0.0988 | No | ||
| 44 | GGT5 | 25506 | -0.378 | -0.0761 | No | ||
| 45 | GPX7 | 25564 | -0.381 | -0.0487 | No | ||
| 46 | GPX3 | 25596 | -0.382 | -0.0203 | No | ||
| 47 | GSTM3 | 26567 | -0.440 | -0.0206 | No | ||
| 48 | GSTM5 | 27452 | -0.515 | -0.0120 | No | ||
| 49 | GSTP1 | 28082 | -0.601 | 0.0122 | No |