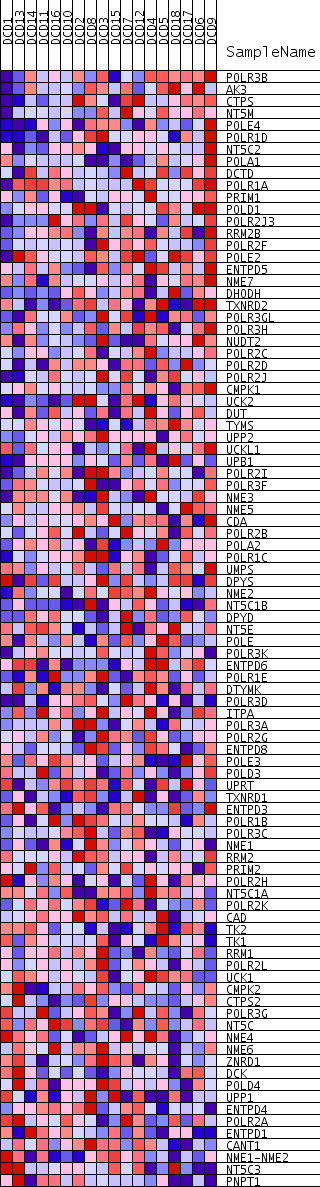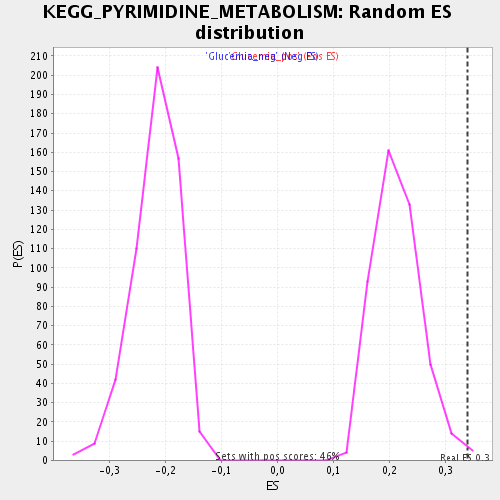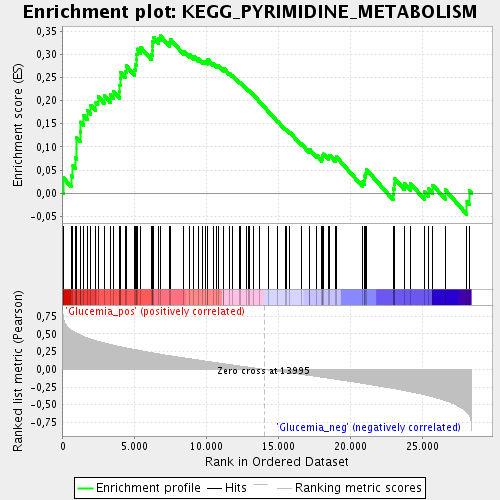
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | DCD_collapsed_to_symbols.DCD.cls#Glucemia |
| Phenotype | DCD.cls#Glucemia |
| Upregulated in class | Glucemia_pos |
| GeneSet | KEGG_PYRIMIDINE_METABOLISM |
| Enrichment Score (ES) | 0.3398547 |
| Normalized Enrichment Score (NES) | 1.597693 |
| Nominal p-value | 0.008695652 |
| FDR q-value | 0.08789882 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | POLR3B | 17 | 0.742 | 0.0333 | Yes | ||
| 2 | AK3 | 583 | 0.549 | 0.0384 | Yes | ||
| 3 | CTPS | 695 | 0.536 | 0.0589 | Yes | ||
| 4 | NT5M | 867 | 0.518 | 0.0765 | Yes | ||
| 5 | POLE4 | 964 | 0.508 | 0.0963 | Yes | ||
| 6 | POLR1D | 965 | 0.508 | 0.1194 | Yes | ||
| 7 | NT5C2 | 1202 | 0.486 | 0.1332 | Yes | ||
| 8 | POLA1 | 1242 | 0.481 | 0.1538 | Yes | ||
| 9 | DCTD | 1455 | 0.461 | 0.1673 | Yes | ||
| 10 | POLR1A | 1689 | 0.445 | 0.1794 | Yes | ||
| 11 | PRIM1 | 1933 | 0.429 | 0.1904 | Yes | ||
| 12 | POLD1 | 2297 | 0.405 | 0.1960 | Yes | ||
| 13 | POLR2J3 | 2439 | 0.395 | 0.2090 | Yes | ||
| 14 | RRM2B | 2863 | 0.373 | 0.2111 | Yes | ||
| 15 | POLR2F | 3283 | 0.353 | 0.2124 | Yes | ||
| 16 | POLE2 | 3494 | 0.343 | 0.2206 | Yes | ||
| 17 | ENTPD5 | 3933 | 0.322 | 0.2199 | Yes | ||
| 18 | NME7 | 3962 | 0.321 | 0.2335 | Yes | ||
| 19 | DHODH | 3981 | 0.320 | 0.2475 | Yes | ||
| 20 | TXNRD2 | 3995 | 0.320 | 0.2616 | Yes | ||
| 21 | POLR3GL | 4333 | 0.306 | 0.2637 | Yes | ||
| 22 | POLR3H | 4391 | 0.304 | 0.2755 | Yes | ||
| 23 | NUDT2 | 5009 | 0.279 | 0.2665 | Yes | ||
| 24 | POLR2C | 5052 | 0.278 | 0.2776 | Yes | ||
| 25 | POLR2D | 5094 | 0.276 | 0.2888 | Yes | ||
| 26 | POLR2J | 5122 | 0.275 | 0.3004 | Yes | ||
| 27 | CMPK1 | 5161 | 0.274 | 0.3115 | Yes | ||
| 28 | UCK2 | 5415 | 0.264 | 0.3146 | Yes | ||
| 29 | DUT | 6159 | 0.236 | 0.2991 | Yes | ||
| 30 | TYMS | 6219 | 0.233 | 0.3077 | Yes | ||
| 31 | UPP2 | 6226 | 0.233 | 0.3181 | Yes | ||
| 32 | UCKL1 | 6235 | 0.233 | 0.3285 | Yes | ||
| 33 | UPB1 | 6302 | 0.231 | 0.3366 | Yes | ||
| 34 | POLR2I | 6677 | 0.218 | 0.3334 | Yes | ||
| 35 | POLR3F | 6771 | 0.214 | 0.3399 | Yes | ||
| 36 | NME3 | 7409 | 0.193 | 0.3262 | No | ||
| 37 | NME5 | 7470 | 0.191 | 0.3327 | No | ||
| 38 | CDA | 8410 | 0.162 | 0.3070 | No | ||
| 39 | POLR2B | 8830 | 0.149 | 0.2990 | No | ||
| 40 | POLA2 | 9104 | 0.141 | 0.2958 | No | ||
| 41 | POLR1C | 9429 | 0.131 | 0.2903 | No | ||
| 42 | UMPS | 9736 | 0.122 | 0.2851 | No | ||
| 43 | DPYS | 9888 | 0.117 | 0.2851 | No | ||
| 44 | NME2 | 10061 | 0.112 | 0.2841 | No | ||
| 45 | NT5C1B | 10070 | 0.111 | 0.2889 | No | ||
| 46 | DPYD | 10444 | 0.100 | 0.2803 | No | ||
| 47 | NT5E | 10674 | 0.094 | 0.2765 | No | ||
| 48 | POLE | 10795 | 0.090 | 0.2763 | No | ||
| 49 | POLR3K | 11174 | 0.079 | 0.2666 | No | ||
| 50 | ENTPD6 | 11195 | 0.079 | 0.2695 | No | ||
| 51 | POLR1E | 11587 | 0.068 | 0.2588 | No | ||
| 52 | DTYMK | 11797 | 0.061 | 0.2542 | No | ||
| 53 | POLR3D | 12293 | 0.047 | 0.2389 | No | ||
| 54 | ITPA | 12349 | 0.046 | 0.2390 | No | ||
| 55 | POLR3A | 12762 | 0.035 | 0.2261 | No | ||
| 56 | POLR2G | 12932 | 0.030 | 0.2215 | No | ||
| 57 | ENTPD8 | 12950 | 0.030 | 0.2222 | No | ||
| 58 | POLE3 | 13272 | 0.020 | 0.2118 | No | ||
| 59 | POLD3 | 13705 | 0.008 | 0.1969 | No | ||
| 60 | UPRT | 14330 | -0.010 | 0.1754 | No | ||
| 61 | TXNRD1 | 14949 | -0.027 | 0.1548 | No | ||
| 62 | ENTPD3 | 15468 | -0.041 | 0.1384 | No | ||
| 63 | POLR1B | 15537 | -0.043 | 0.1380 | No | ||
| 64 | POLR3C | 15788 | -0.050 | 0.1315 | No | ||
| 65 | NME1 | 16585 | -0.072 | 0.1067 | No | ||
| 66 | RRM2 | 17130 | -0.087 | 0.0914 | No | ||
| 67 | PRIM2 | 17173 | -0.088 | 0.0940 | No | ||
| 68 | POLR2H | 17650 | -0.101 | 0.0817 | No | ||
| 69 | NT5C1A | 18018 | -0.111 | 0.0739 | No | ||
| 70 | POLR2K | 18038 | -0.112 | 0.0783 | No | ||
| 71 | CAD | 18073 | -0.113 | 0.0822 | No | ||
| 72 | TK2 | 18154 | -0.115 | 0.0846 | No | ||
| 73 | TK1 | 18483 | -0.125 | 0.0788 | No | ||
| 74 | RRM1 | 18555 | -0.127 | 0.0820 | No | ||
| 75 | POLR2L | 18959 | -0.140 | 0.0742 | No | ||
| 76 | UCK1 | 19004 | -0.141 | 0.0791 | No | ||
| 77 | CMPK2 | 20813 | -0.197 | 0.0243 | No | ||
| 78 | CTPS2 | 20964 | -0.202 | 0.0282 | No | ||
| 79 | POLR3G | 20973 | -0.202 | 0.0371 | No | ||
| 80 | NT5C | 21080 | -0.206 | 0.0428 | No | ||
| 81 | NME4 | 21102 | -0.207 | 0.0515 | No | ||
| 82 | NME6 | 22972 | -0.270 | -0.0022 | No | ||
| 83 | ZNRD1 | 22988 | -0.270 | 0.0096 | No | ||
| 84 | DCK | 23052 | -0.273 | 0.0198 | No | ||
| 85 | POLD4 | 23096 | -0.274 | 0.0308 | No | ||
| 86 | UPP1 | 23756 | -0.300 | 0.0213 | No | ||
| 87 | ENTPD4 | 24186 | -0.317 | 0.0206 | No | ||
| 88 | POLR2A | 25142 | -0.359 | 0.0032 | No | ||
| 89 | ENTPD1 | 25447 | -0.376 | 0.0096 | No | ||
| 90 | CANT1 | 25730 | -0.390 | 0.0175 | No | ||
| 91 | NME1-NME2 | 26589 | -0.441 | 0.0073 | No | ||
| 92 | NT5C3 | 28108 | -0.606 | -0.0186 | No | ||
| 93 | PNPT1 | 28274 | -0.655 | 0.0054 | No |