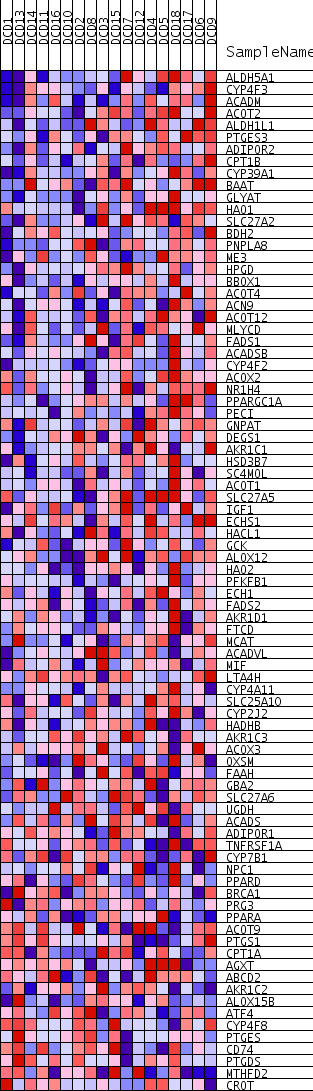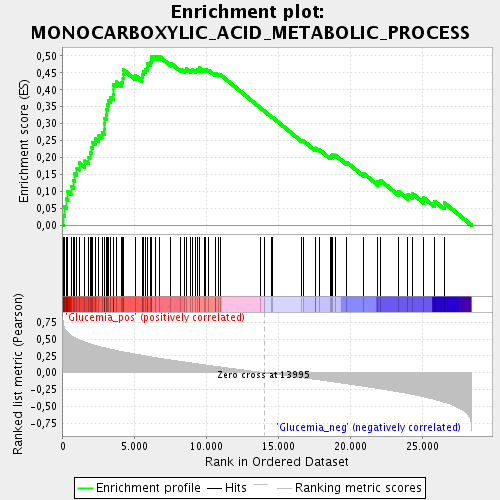
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | DCD_collapsed_to_symbols.DCD.cls#Glucemia |
| Phenotype | DCD.cls#Glucemia |
| Upregulated in class | Glucemia_pos |
| GeneSet | MONOCARBOXYLIC_ACID_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.4989176 |
| Normalized Enrichment Score (NES) | 2.2738013 |
| Nominal p-value | 0.0 |
| FDR q-value | 1.1785148E-4 |
| FWER p-Value | 0.0030 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ALDH5A1 | 46 | 0.706 | 0.0288 | Yes | ||
| 2 | CYP4F3 | 120 | 0.660 | 0.0547 | Yes | ||
| 3 | ACADM | 225 | 0.625 | 0.0780 | Yes | ||
| 4 | ACOT2 | 339 | 0.597 | 0.0997 | Yes | ||
| 5 | ALDH1L1 | 574 | 0.551 | 0.1152 | Yes | ||
| 6 | PTGES3 | 721 | 0.533 | 0.1330 | Yes | ||
| 7 | ADIPOR2 | 796 | 0.526 | 0.1531 | Yes | ||
| 8 | CPT1B | 972 | 0.507 | 0.1688 | Yes | ||
| 9 | CYP39A1 | 1125 | 0.492 | 0.1846 | Yes | ||
| 10 | BAAT | 1528 | 0.456 | 0.1901 | Yes | ||
| 11 | GLYAT | 1764 | 0.439 | 0.2007 | Yes | ||
| 12 | HAO1 | 1882 | 0.431 | 0.2152 | Yes | ||
| 13 | SLC27A2 | 2006 | 0.423 | 0.2291 | Yes | ||
| 14 | BDH2 | 2050 | 0.420 | 0.2457 | Yes | ||
| 15 | PNPLA8 | 2239 | 0.408 | 0.2567 | Yes | ||
| 16 | ME3 | 2498 | 0.392 | 0.2645 | Yes | ||
| 17 | HPGD | 2717 | 0.380 | 0.2732 | Yes | ||
| 18 | BBOX1 | 2869 | 0.373 | 0.2840 | Yes | ||
| 19 | ACOT4 | 2878 | 0.373 | 0.2998 | Yes | ||
| 20 | ACN9 | 2894 | 0.372 | 0.3153 | Yes | ||
| 21 | ACOT12 | 3001 | 0.366 | 0.3273 | Yes | ||
| 22 | MLYCD | 3047 | 0.364 | 0.3414 | Yes | ||
| 23 | FADS1 | 3086 | 0.362 | 0.3557 | Yes | ||
| 24 | ACADSB | 3185 | 0.357 | 0.3677 | Yes | ||
| 25 | CYP4F2 | 3299 | 0.351 | 0.3788 | Yes | ||
| 26 | ACOX2 | 3523 | 0.341 | 0.3857 | Yes | ||
| 27 | NR1H4 | 3533 | 0.341 | 0.4001 | Yes | ||
| 28 | PPARGC1A | 3535 | 0.341 | 0.4147 | Yes | ||
| 29 | PECI | 3694 | 0.333 | 0.4235 | Yes | ||
| 30 | GNPAT | 4100 | 0.315 | 0.4228 | Yes | ||
| 31 | DEGS1 | 4176 | 0.313 | 0.4337 | Yes | ||
| 32 | AKR1C1 | 4205 | 0.311 | 0.4461 | Yes | ||
| 33 | HSD3B7 | 4212 | 0.311 | 0.4593 | Yes | ||
| 34 | SC4MOL | 5028 | 0.278 | 0.4426 | Yes | ||
| 35 | ACOT1 | 5501 | 0.261 | 0.4371 | Yes | ||
| 36 | SLC27A5 | 5544 | 0.259 | 0.4468 | Yes | ||
| 37 | IGF1 | 5626 | 0.256 | 0.4550 | Yes | ||
| 38 | ECHS1 | 5756 | 0.250 | 0.4613 | Yes | ||
| 39 | HACL1 | 5889 | 0.245 | 0.4672 | Yes | ||
| 40 | GCK | 5897 | 0.245 | 0.4775 | Yes | ||
| 41 | ALOX12 | 6072 | 0.239 | 0.4817 | Yes | ||
| 42 | HAO2 | 6139 | 0.236 | 0.4895 | Yes | ||
| 43 | PFKFB1 | 6164 | 0.235 | 0.4988 | Yes | ||
| 44 | ECH1 | 6439 | 0.226 | 0.4989 | Yes | ||
| 45 | FADS2 | 6707 | 0.216 | 0.4988 | No | ||
| 46 | AKR1D1 | 7513 | 0.189 | 0.4786 | No | ||
| 47 | FTCD | 8208 | 0.169 | 0.4614 | No | ||
| 48 | MCAT | 8489 | 0.160 | 0.4584 | No | ||
| 49 | ACADVL | 8573 | 0.157 | 0.4622 | No | ||
| 50 | MIF | 8895 | 0.147 | 0.4573 | No | ||
| 51 | LTA4H | 9005 | 0.144 | 0.4596 | No | ||
| 52 | CYP4A11 | 9224 | 0.137 | 0.4579 | No | ||
| 53 | SLC25A10 | 9341 | 0.134 | 0.4596 | No | ||
| 54 | CYP2J2 | 9476 | 0.129 | 0.4604 | No | ||
| 55 | HADHB | 9488 | 0.129 | 0.4656 | No | ||
| 56 | AKR1C3 | 9851 | 0.118 | 0.4579 | No | ||
| 57 | ACOX3 | 9918 | 0.116 | 0.4606 | No | ||
| 58 | OXSM | 10113 | 0.110 | 0.4585 | No | ||
| 59 | FAAH | 10594 | 0.096 | 0.4457 | No | ||
| 60 | GBA2 | 10641 | 0.094 | 0.4482 | No | ||
| 61 | SLC27A6 | 10825 | 0.089 | 0.4455 | No | ||
| 62 | UGDH | 10932 | 0.086 | 0.4455 | No | ||
| 63 | ACADS | 13736 | 0.007 | 0.3469 | No | ||
| 64 | ADIPOR1 | 14031 | -0.001 | 0.3366 | No | ||
| 65 | TNFRSF1A | 14539 | -0.016 | 0.3194 | No | ||
| 66 | CYP7B1 | 14595 | -0.017 | 0.3182 | No | ||
| 67 | NPC1 | 16571 | -0.072 | 0.2516 | No | ||
| 68 | PPARD | 16724 | -0.076 | 0.2495 | No | ||
| 69 | BRCA1 | 17535 | -0.098 | 0.2251 | No | ||
| 70 | PRG3 | 17564 | -0.099 | 0.2284 | No | ||
| 71 | PPARA | 17824 | -0.106 | 0.2238 | No | ||
| 72 | ACOT9 | 18604 | -0.129 | 0.2019 | No | ||
| 73 | PTGS1 | 18657 | -0.130 | 0.2057 | No | ||
| 74 | CPT1A | 18755 | -0.133 | 0.2080 | No | ||
| 75 | AGXT | 18933 | -0.139 | 0.2077 | No | ||
| 76 | ABCD2 | 19723 | -0.163 | 0.1870 | No | ||
| 77 | AKR1C2 | 20922 | -0.201 | 0.1533 | No | ||
| 78 | ALOX15B | 21905 | -0.233 | 0.1287 | No | ||
| 79 | ATF4 | 22087 | -0.239 | 0.1327 | No | ||
| 80 | CYP4F8 | 23337 | -0.284 | 0.1008 | No | ||
| 81 | PTGES | 24003 | -0.310 | 0.0907 | No | ||
| 82 | CD74 | 24291 | -0.322 | 0.0945 | No | ||
| 83 | PTGDS | 25109 | -0.357 | 0.0811 | No | ||
| 84 | MTHFD2 | 25858 | -0.397 | 0.0718 | No | ||
| 85 | CROT | 26517 | -0.437 | 0.0674 | No |