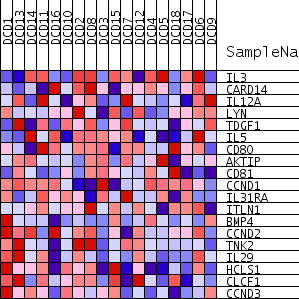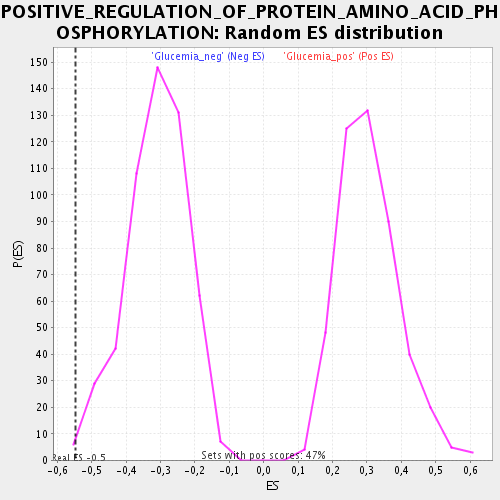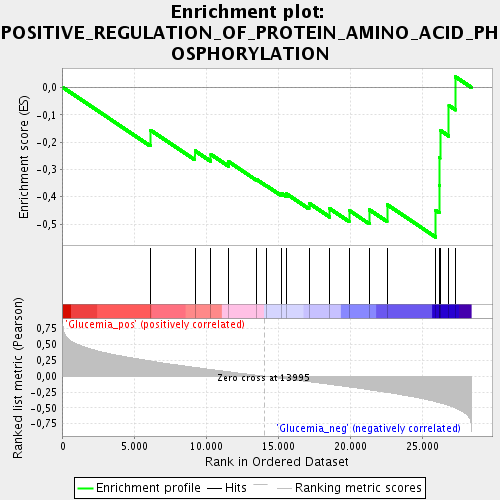
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | DCD_collapsed_to_symbols.DCD.cls#Glucemia |
| Phenotype | DCD.cls#Glucemia |
| Upregulated in class | Glucemia_neg |
| GeneSet | POSITIVE_REGULATION_OF_PROTEIN_AMINO_ACID_PHOSPHORYLATION |
| Enrichment Score (ES) | -0.5482559 |
| Normalized Enrichment Score (NES) | -1.7601237 |
| Nominal p-value | 0.0075046904 |
| FDR q-value | 0.16615984 |
| FWER p-Value | 0.971 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IL3 | 6077 | 0.239 | -0.1559 | No | ||
| 2 | CARD14 | 9186 | 0.138 | -0.2316 | No | ||
| 3 | IL12A | 10297 | 0.104 | -0.2454 | No | ||
| 4 | LYN | 11511 | 0.070 | -0.2711 | No | ||
| 5 | TDGF1 | 13472 | 0.015 | -0.3365 | No | ||
| 6 | IL5 | 14158 | -0.005 | -0.3594 | No | ||
| 7 | CD80 | 15229 | -0.035 | -0.3887 | No | ||
| 8 | AKTIP | 15532 | -0.043 | -0.3888 | No | ||
| 9 | CD81 | 17119 | -0.086 | -0.4237 | No | ||
| 10 | CCND1 | 18562 | -0.127 | -0.4435 | No | ||
| 11 | IL31RA | 19907 | -0.169 | -0.4498 | No | ||
| 12 | ITLN1 | 21324 | -0.214 | -0.4477 | No | ||
| 13 | BMP4 | 22546 | -0.255 | -0.4287 | No | ||
| 14 | CCND2 | 25943 | -0.402 | -0.4506 | Yes | ||
| 15 | TNK2 | 26170 | -0.416 | -0.3575 | Yes | ||
| 16 | IL29 | 26202 | -0.418 | -0.2570 | Yes | ||
| 17 | HCLS1 | 26232 | -0.419 | -0.1561 | Yes | ||
| 18 | CLCF1 | 26855 | -0.461 | -0.0661 | Yes | ||
| 19 | CCND3 | 27316 | -0.500 | 0.0391 | Yes |