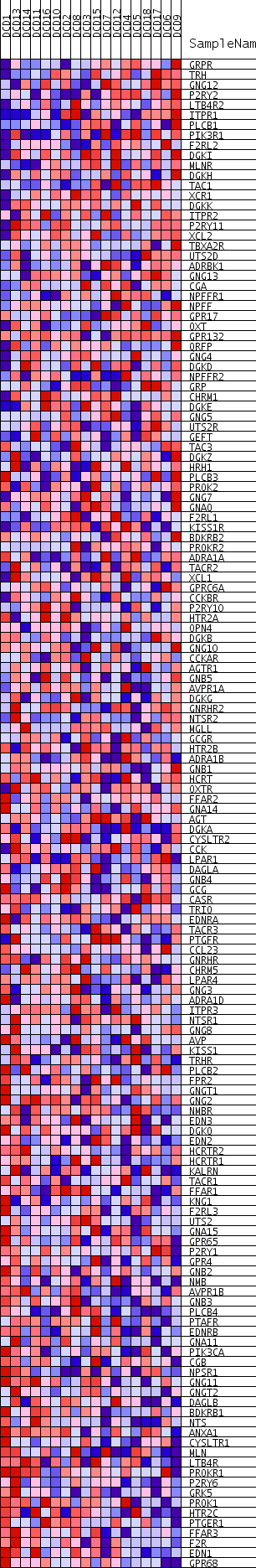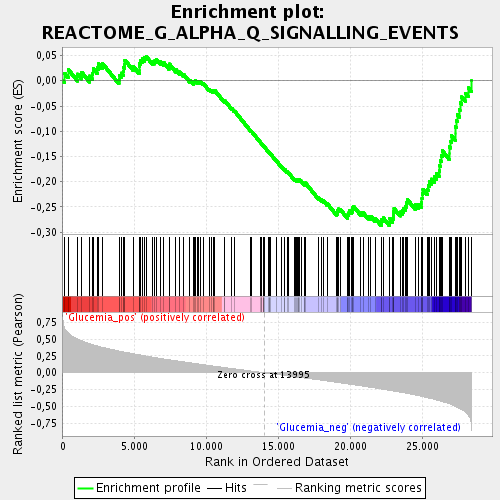
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | DCD_collapsed_to_symbols.DCD.cls#Glucemia |
| Phenotype | DCD.cls#Glucemia |
| Upregulated in class | Glucemia_neg |
| GeneSet | REACTOME_G_ALPHA_Q_SIGNALLING_EVENTS |
| Enrichment Score (ES) | -0.28752154 |
| Normalized Enrichment Score (NES) | -1.4126147 |
| Nominal p-value | 0.014625229 |
| FDR q-value | 0.5727984 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GRPR | 100 | 0.670 | 0.0152 | No | ||
| 2 | TRH | 361 | 0.592 | 0.0226 | No | ||
| 3 | GNG12 | 1020 | 0.502 | 0.0135 | No | ||
| 4 | P2RY2 | 1304 | 0.476 | 0.0168 | No | ||
| 5 | LTB4R2 | 1854 | 0.433 | 0.0095 | No | ||
| 6 | ITPR1 | 2045 | 0.421 | 0.0146 | No | ||
| 7 | PLCB1 | 2090 | 0.418 | 0.0247 | No | ||
| 8 | PIK3R1 | 2381 | 0.399 | 0.0256 | No | ||
| 9 | F2RL2 | 2469 | 0.393 | 0.0336 | No | ||
| 10 | DGKI | 2743 | 0.379 | 0.0346 | No | ||
| 11 | MLNR | 3902 | 0.324 | 0.0027 | No | ||
| 12 | DGKH | 3941 | 0.322 | 0.0103 | No | ||
| 13 | TAC1 | 4050 | 0.318 | 0.0154 | No | ||
| 14 | XCR1 | 4191 | 0.312 | 0.0192 | No | ||
| 15 | DGKK | 4216 | 0.311 | 0.0271 | No | ||
| 16 | ITPR2 | 4298 | 0.307 | 0.0328 | No | ||
| 17 | P2RY11 | 4308 | 0.307 | 0.0411 | No | ||
| 18 | XCL2 | 4912 | 0.283 | 0.0277 | No | ||
| 19 | TBXA2R | 5313 | 0.268 | 0.0210 | No | ||
| 20 | UTS2D | 5318 | 0.267 | 0.0284 | No | ||
| 21 | ADRBK1 | 5345 | 0.266 | 0.0349 | No | ||
| 22 | GNG13 | 5408 | 0.264 | 0.0401 | No | ||
| 23 | CGA | 5509 | 0.260 | 0.0439 | No | ||
| 24 | NPFFR1 | 5661 | 0.254 | 0.0457 | No | ||
| 25 | NPFF | 5784 | 0.249 | 0.0484 | No | ||
| 26 | GPR17 | 6233 | 0.233 | 0.0390 | No | ||
| 27 | OXT | 6400 | 0.227 | 0.0395 | No | ||
| 28 | GPR132 | 6507 | 0.224 | 0.0421 | No | ||
| 29 | QRFP | 6796 | 0.214 | 0.0379 | No | ||
| 30 | GNG4 | 7004 | 0.206 | 0.0363 | No | ||
| 31 | DGKD | 7381 | 0.194 | 0.0284 | No | ||
| 32 | NPFFR2 | 7397 | 0.193 | 0.0333 | No | ||
| 33 | GRP | 7853 | 0.179 | 0.0222 | No | ||
| 34 | CHRM1 | 8121 | 0.171 | 0.0176 | No | ||
| 35 | DGKE | 8379 | 0.163 | 0.0131 | No | ||
| 36 | GNG5 | 8831 | 0.149 | 0.0013 | No | ||
| 37 | UTS2R | 9077 | 0.141 | -0.0034 | No | ||
| 38 | GEFT | 9121 | 0.140 | -0.0010 | No | ||
| 39 | TAC3 | 9188 | 0.138 | 0.0005 | No | ||
| 40 | DGKZ | 9354 | 0.134 | -0.0016 | No | ||
| 41 | HRH1 | 9464 | 0.130 | -0.0018 | No | ||
| 42 | PLCB3 | 9549 | 0.127 | -0.0012 | No | ||
| 43 | PROK2 | 9753 | 0.121 | -0.0050 | No | ||
| 44 | GNG7 | 10166 | 0.108 | -0.0165 | No | ||
| 45 | GNAQ | 10300 | 0.104 | -0.0183 | No | ||
| 46 | F2RL1 | 10440 | 0.100 | -0.0204 | No | ||
| 47 | KISS1R | 10507 | 0.098 | -0.0200 | No | ||
| 48 | BDKRB2 | 10560 | 0.097 | -0.0191 | No | ||
| 49 | PROKR2 | 11257 | 0.077 | -0.0416 | No | ||
| 50 | ADRA1A | 11270 | 0.076 | -0.0399 | No | ||
| 51 | TACR2 | 11736 | 0.063 | -0.0545 | No | ||
| 52 | XCL1 | 11933 | 0.058 | -0.0599 | No | ||
| 53 | GPRC6A | 13081 | 0.026 | -0.0997 | No | ||
| 54 | CCKBR | 13120 | 0.025 | -0.1004 | No | ||
| 55 | P2RY10 | 13753 | 0.007 | -0.1225 | No | ||
| 56 | HTR2A | 13800 | 0.005 | -0.1240 | No | ||
| 57 | OPN4 | 13817 | 0.005 | -0.1244 | No | ||
| 58 | DGKB | 13925 | 0.002 | -0.1282 | No | ||
| 59 | GNG10 | 13957 | 0.001 | -0.1292 | No | ||
| 60 | CCKAR | 13964 | 0.001 | -0.1294 | No | ||
| 61 | AGTR1 | 14025 | -0.001 | -0.1315 | No | ||
| 62 | GNB5 | 14269 | -0.008 | -0.1399 | No | ||
| 63 | AVPR1A | 14338 | -0.010 | -0.1420 | No | ||
| 64 | DGKG | 14382 | -0.011 | -0.1432 | No | ||
| 65 | GNRHR2 | 14406 | -0.011 | -0.1437 | No | ||
| 66 | NTSR2 | 14883 | -0.026 | -0.1599 | No | ||
| 67 | MGLL | 15230 | -0.035 | -0.1711 | No | ||
| 68 | GCGR | 15433 | -0.041 | -0.1771 | No | ||
| 69 | HTR2B | 15444 | -0.041 | -0.1763 | No | ||
| 70 | ADRA1B | 15588 | -0.045 | -0.1801 | No | ||
| 71 | GNB1 | 15708 | -0.048 | -0.1830 | No | ||
| 72 | HCRT | 16104 | -0.059 | -0.1953 | No | ||
| 73 | OXTR | 16160 | -0.061 | -0.1956 | No | ||
| 74 | FFAR2 | 16267 | -0.063 | -0.1975 | No | ||
| 75 | GNA14 | 16307 | -0.064 | -0.1971 | No | ||
| 76 | AGT | 16352 | -0.066 | -0.1968 | No | ||
| 77 | DGKA | 16364 | -0.066 | -0.1953 | No | ||
| 78 | CYSLTR2 | 16424 | -0.068 | -0.1955 | No | ||
| 79 | CCK | 16566 | -0.072 | -0.1985 | No | ||
| 80 | LPAR1 | 16803 | -0.078 | -0.2047 | No | ||
| 81 | DAGLA | 16817 | -0.078 | -0.2030 | No | ||
| 82 | GNB4 | 16863 | -0.079 | -0.2023 | No | ||
| 83 | GCG | 17777 | -0.104 | -0.2317 | No | ||
| 84 | CASR | 17969 | -0.110 | -0.2354 | No | ||
| 85 | TRIO | 18124 | -0.114 | -0.2376 | No | ||
| 86 | EDNRA | 18388 | -0.122 | -0.2435 | No | ||
| 87 | TACR3 | 19032 | -0.142 | -0.2622 | No | ||
| 88 | PTGFR | 19093 | -0.144 | -0.2603 | No | ||
| 89 | CCL23 | 19111 | -0.145 | -0.2569 | No | ||
| 90 | GNRHR | 19144 | -0.146 | -0.2539 | No | ||
| 91 | CHRM5 | 19294 | -0.151 | -0.2549 | No | ||
| 92 | LPAR4 | 19799 | -0.165 | -0.2681 | No | ||
| 93 | GNG3 | 19827 | -0.166 | -0.2644 | No | ||
| 94 | ADRA1D | 19888 | -0.168 | -0.2618 | No | ||
| 95 | ITPR3 | 19903 | -0.169 | -0.2576 | No | ||
| 96 | NTSR1 | 20082 | -0.174 | -0.2590 | No | ||
| 97 | GNG8 | 20117 | -0.175 | -0.2553 | No | ||
| 98 | AVP | 20155 | -0.176 | -0.2517 | No | ||
| 99 | KISS1 | 20236 | -0.178 | -0.2495 | No | ||
| 100 | TRHR | 20733 | -0.195 | -0.2616 | No | ||
| 101 | PLCB2 | 20894 | -0.200 | -0.2617 | No | ||
| 102 | FPR2 | 21243 | -0.211 | -0.2681 | No | ||
| 103 | GNGT1 | 21427 | -0.218 | -0.2684 | No | ||
| 104 | GNG2 | 21725 | -0.227 | -0.2726 | No | ||
| 105 | NMBR | 22148 | -0.242 | -0.2807 | Yes | ||
| 106 | EDN3 | 22185 | -0.243 | -0.2752 | Yes | ||
| 107 | DGKQ | 22278 | -0.246 | -0.2716 | Yes | ||
| 108 | EDN2 | 22730 | -0.261 | -0.2802 | Yes | ||
| 109 | HCRTR2 | 22744 | -0.262 | -0.2733 | Yes | ||
| 110 | HCRTR1 | 22960 | -0.270 | -0.2734 | Yes | ||
| 111 | KALRN | 22982 | -0.270 | -0.2665 | Yes | ||
| 112 | TACR1 | 23002 | -0.271 | -0.2596 | Yes | ||
| 113 | FFAR1 | 23029 | -0.272 | -0.2529 | Yes | ||
| 114 | KNG1 | 23455 | -0.289 | -0.2599 | Yes | ||
| 115 | F2RL3 | 23620 | -0.295 | -0.2574 | Yes | ||
| 116 | UTS2 | 23716 | -0.299 | -0.2524 | Yes | ||
| 117 | GNA15 | 23849 | -0.303 | -0.2486 | Yes | ||
| 118 | GPR65 | 23899 | -0.305 | -0.2418 | Yes | ||
| 119 | P2RY1 | 23940 | -0.307 | -0.2346 | Yes | ||
| 120 | GPR4 | 24519 | -0.332 | -0.2457 | Yes | ||
| 121 | GNB2 | 24750 | -0.340 | -0.2443 | Yes | ||
| 122 | NMB | 24951 | -0.349 | -0.2416 | Yes | ||
| 123 | AVPR1B | 24975 | -0.350 | -0.2326 | Yes | ||
| 124 | GNB3 | 24989 | -0.351 | -0.2233 | Yes | ||
| 125 | PLCB4 | 25044 | -0.354 | -0.2153 | Yes | ||
| 126 | PTAFR | 25341 | -0.370 | -0.2154 | Yes | ||
| 127 | EDNRB | 25415 | -0.374 | -0.2075 | Yes | ||
| 128 | GNA11 | 25497 | -0.378 | -0.1997 | Yes | ||
| 129 | PIK3CA | 25657 | -0.385 | -0.1946 | Yes | ||
| 130 | CGB | 25839 | -0.396 | -0.1899 | Yes | ||
| 131 | NPSR1 | 25988 | -0.405 | -0.1838 | Yes | ||
| 132 | GNG11 | 26171 | -0.416 | -0.1785 | Yes | ||
| 133 | GNGT2 | 26210 | -0.418 | -0.1682 | Yes | ||
| 134 | DAGLB | 26255 | -0.421 | -0.1579 | Yes | ||
| 135 | BDKRB1 | 26330 | -0.426 | -0.1486 | Yes | ||
| 136 | NTS | 26389 | -0.429 | -0.1386 | Yes | ||
| 137 | ANXA1 | 26904 | -0.464 | -0.1438 | Yes | ||
| 138 | CYSLTR1 | 26906 | -0.464 | -0.1309 | Yes | ||
| 139 | MLN | 26976 | -0.469 | -0.1202 | Yes | ||
| 140 | LTB4R | 27036 | -0.476 | -0.1089 | Yes | ||
| 141 | PROKR1 | 27287 | -0.497 | -0.1038 | Yes | ||
| 142 | P2RY6 | 27288 | -0.497 | -0.0899 | Yes | ||
| 143 | GRK5 | 27382 | -0.508 | -0.0790 | Yes | ||
| 144 | PROK1 | 27427 | -0.513 | -0.0661 | Yes | ||
| 145 | HTR2C | 27594 | -0.531 | -0.0571 | Yes | ||
| 146 | PTGER1 | 27628 | -0.535 | -0.0433 | Yes | ||
| 147 | FFAR3 | 27742 | -0.547 | -0.0320 | Yes | ||
| 148 | F2R | 28002 | -0.586 | -0.0247 | Yes | ||
| 149 | EDN1 | 28201 | -0.631 | -0.0140 | Yes | ||
| 150 | GPR68 | 28421 | -0.785 | 0.0002 | Yes |