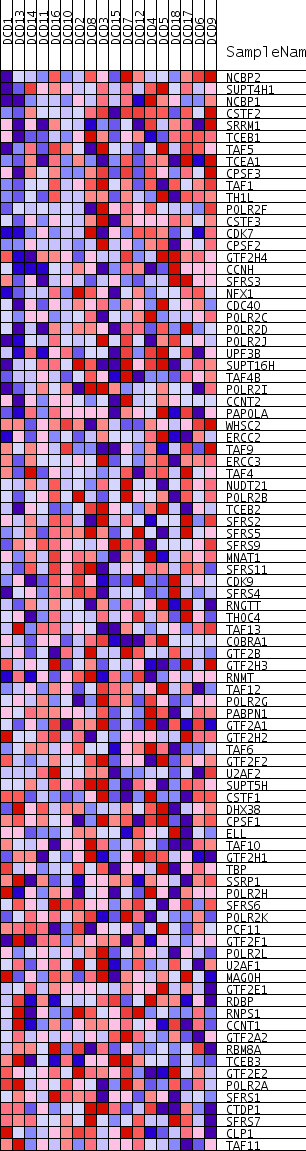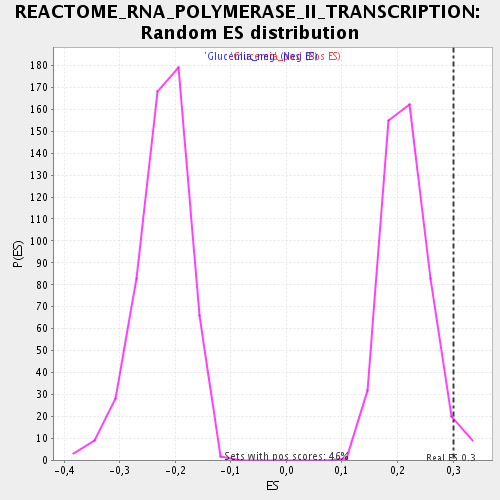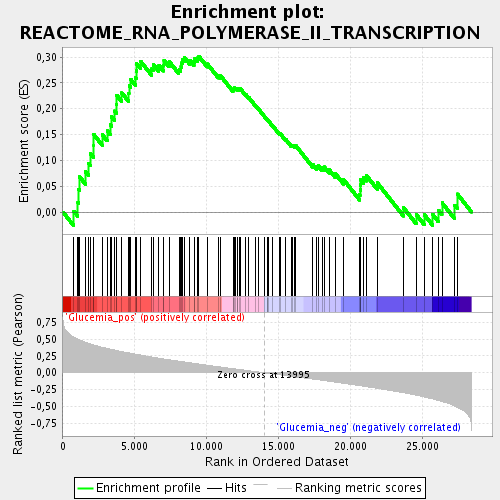
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | DCD_collapsed_to_symbols.DCD.cls#Glucemia |
| Phenotype | DCD.cls#Glucemia |
| Upregulated in class | Glucemia_pos |
| GeneSet | REACTOME_RNA_POLYMERASE_II_TRANSCRIPTION |
| Enrichment Score (ES) | 0.30134934 |
| Normalized Enrichment Score (NES) | 1.4029707 |
| Nominal p-value | 0.032467533 |
| FDR q-value | 0.2008962 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | NCBP2 | 760 | 0.529 | 0.0018 | Yes | ||
| 2 | SUPT4H1 | 1040 | 0.501 | 0.0190 | Yes | ||
| 3 | NCBP1 | 1093 | 0.496 | 0.0440 | Yes | ||
| 4 | CSTF2 | 1158 | 0.489 | 0.0682 | Yes | ||
| 5 | SRRM1 | 1571 | 0.452 | 0.0781 | Yes | ||
| 6 | TCEB1 | 1781 | 0.438 | 0.0944 | Yes | ||
| 7 | TAF5 | 1908 | 0.430 | 0.1133 | Yes | ||
| 8 | TCEA1 | 2114 | 0.416 | 0.1285 | Yes | ||
| 9 | CPSF3 | 2120 | 0.416 | 0.1508 | Yes | ||
| 10 | TAF1 | 2729 | 0.380 | 0.1499 | Yes | ||
| 11 | TH1L | 3080 | 0.362 | 0.1572 | Yes | ||
| 12 | POLR2F | 3283 | 0.353 | 0.1691 | Yes | ||
| 13 | CSTF3 | 3379 | 0.348 | 0.1845 | Yes | ||
| 14 | CDK7 | 3584 | 0.338 | 0.1956 | Yes | ||
| 15 | CPSF2 | 3717 | 0.332 | 0.2089 | Yes | ||
| 16 | GTF2H4 | 3735 | 0.331 | 0.2263 | Yes | ||
| 17 | CCNH | 4095 | 0.315 | 0.2306 | Yes | ||
| 18 | SFRS3 | 4559 | 0.298 | 0.2304 | Yes | ||
| 19 | NFX1 | 4598 | 0.296 | 0.2451 | Yes | ||
| 20 | CDC40 | 4719 | 0.291 | 0.2566 | Yes | ||
| 21 | POLR2C | 5052 | 0.278 | 0.2599 | Yes | ||
| 22 | POLR2D | 5094 | 0.276 | 0.2734 | Yes | ||
| 23 | POLR2J | 5122 | 0.275 | 0.2873 | Yes | ||
| 24 | UPF3B | 5411 | 0.264 | 0.2914 | Yes | ||
| 25 | SUPT16H | 6178 | 0.235 | 0.2771 | Yes | ||
| 26 | TAF4B | 6273 | 0.232 | 0.2863 | Yes | ||
| 27 | POLR2I | 6677 | 0.218 | 0.2838 | Yes | ||
| 28 | CCNT2 | 6986 | 0.207 | 0.2841 | Yes | ||
| 29 | PAPOLA | 7027 | 0.205 | 0.2938 | Yes | ||
| 30 | WHSC2 | 7386 | 0.194 | 0.2916 | Yes | ||
| 31 | ERCC2 | 8073 | 0.172 | 0.2767 | Yes | ||
| 32 | TAF9 | 8210 | 0.169 | 0.2810 | Yes | ||
| 33 | ERCC3 | 8251 | 0.167 | 0.2887 | Yes | ||
| 34 | TAF4 | 8298 | 0.165 | 0.2960 | Yes | ||
| 35 | NUDT21 | 8451 | 0.161 | 0.2993 | Yes | ||
| 36 | POLR2B | 8830 | 0.149 | 0.2940 | Yes | ||
| 37 | TCEB2 | 9120 | 0.140 | 0.2914 | Yes | ||
| 38 | SFRS2 | 9177 | 0.139 | 0.2969 | Yes | ||
| 39 | SFRS5 | 9345 | 0.134 | 0.2983 | Yes | ||
| 40 | SFRS9 | 9458 | 0.130 | 0.3013 | Yes | ||
| 41 | MNAT1 | 10044 | 0.112 | 0.2868 | No | ||
| 42 | SFRS11 | 10820 | 0.089 | 0.2642 | No | ||
| 43 | CDK9 | 10967 | 0.085 | 0.2637 | No | ||
| 44 | SFRS4 | 11833 | 0.060 | 0.2364 | No | ||
| 45 | RNGTT | 11869 | 0.059 | 0.2384 | No | ||
| 46 | THOC4 | 11902 | 0.059 | 0.2404 | No | ||
| 47 | TAF13 | 11999 | 0.056 | 0.2401 | No | ||
| 48 | COBRA1 | 12115 | 0.052 | 0.2388 | No | ||
| 49 | GTF2B | 12159 | 0.051 | 0.2401 | No | ||
| 50 | GTF2H3 | 12299 | 0.047 | 0.2377 | No | ||
| 51 | RNMT | 12344 | 0.046 | 0.2386 | No | ||
| 52 | TAF12 | 12708 | 0.036 | 0.2278 | No | ||
| 53 | POLR2G | 12932 | 0.030 | 0.2216 | No | ||
| 54 | PABPN1 | 13379 | 0.018 | 0.2068 | No | ||
| 55 | GTF2A1 | 13589 | 0.011 | 0.2000 | No | ||
| 56 | GTF2H2 | 14006 | -0.000 | 0.1853 | No | ||
| 57 | TAF6 | 14209 | -0.006 | 0.1786 | No | ||
| 58 | GTF2F2 | 14255 | -0.007 | 0.1774 | No | ||
| 59 | U2AF2 | 14287 | -0.009 | 0.1767 | No | ||
| 60 | SUPT5H | 14567 | -0.016 | 0.1678 | No | ||
| 61 | CSTF1 | 15050 | -0.030 | 0.1524 | No | ||
| 62 | DHX38 | 15120 | -0.032 | 0.1517 | No | ||
| 63 | CPSF1 | 15485 | -0.042 | 0.1411 | No | ||
| 64 | ELL | 15915 | -0.054 | 0.1288 | No | ||
| 65 | TAF10 | 15974 | -0.055 | 0.1298 | No | ||
| 66 | GTF2H1 | 16137 | -0.060 | 0.1273 | No | ||
| 67 | TBP | 16204 | -0.062 | 0.1283 | No | ||
| 68 | SSRP1 | 17377 | -0.094 | 0.0920 | No | ||
| 69 | POLR2H | 17650 | -0.101 | 0.0879 | No | ||
| 70 | SFRS6 | 17755 | -0.104 | 0.0898 | No | ||
| 71 | POLR2K | 18038 | -0.112 | 0.0859 | No | ||
| 72 | PCF11 | 18174 | -0.115 | 0.0874 | No | ||
| 73 | GTF2F1 | 18516 | -0.126 | 0.0822 | No | ||
| 74 | POLR2L | 18959 | -0.140 | 0.0741 | No | ||
| 75 | U2AF1 | 19530 | -0.158 | 0.0625 | No | ||
| 76 | MAGOH | 20648 | -0.192 | 0.0335 | No | ||
| 77 | GTF2E1 | 20671 | -0.193 | 0.0431 | No | ||
| 78 | RDBP | 20700 | -0.194 | 0.0526 | No | ||
| 79 | RNPS1 | 20721 | -0.194 | 0.0624 | No | ||
| 80 | CCNT1 | 20910 | -0.200 | 0.0666 | No | ||
| 81 | GTF2A2 | 21098 | -0.207 | 0.0712 | No | ||
| 82 | RBM8A | 21864 | -0.232 | 0.0567 | No | ||
| 83 | TCEB3 | 23692 | -0.298 | 0.0084 | No | ||
| 84 | GTF2E2 | 24583 | -0.334 | -0.0050 | No | ||
| 85 | POLR2A | 25142 | -0.359 | -0.0053 | No | ||
| 86 | SFRS1 | 25719 | -0.389 | -0.0046 | No | ||
| 87 | CTDP1 | 26125 | -0.414 | 0.0035 | No | ||
| 88 | SFRS7 | 26372 | -0.428 | 0.0179 | No | ||
| 89 | CLP1 | 27252 | -0.494 | 0.0136 | No | ||
| 90 | TAF11 | 27435 | -0.514 | 0.0350 | No |