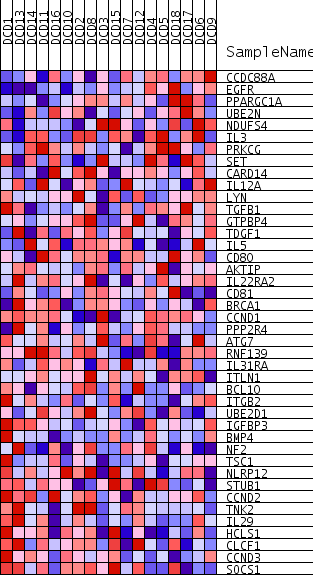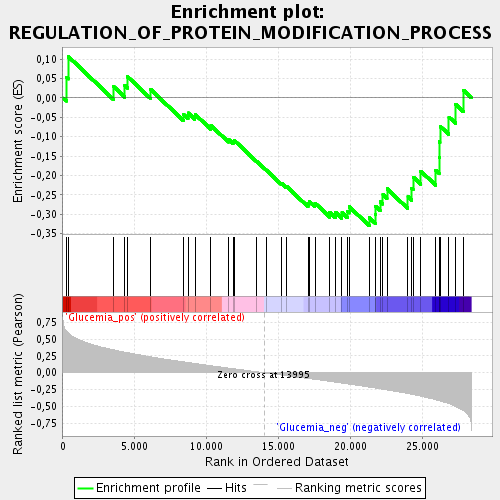
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | DCD_collapsed_to_symbols.DCD.cls#Glucemia |
| Phenotype | DCD.cls#Glucemia |
| Upregulated in class | Glucemia_neg |
| GeneSet | REGULATION_OF_PROTEIN_MODIFICATION_PROCESS |
| Enrichment Score (ES) | -0.3297392 |
| Normalized Enrichment Score (NES) | -1.2893705 |
| Nominal p-value | 0.124087594 |
| FDR q-value | 0.6630418 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CCDC88A | 226 | 0.624 | 0.0535 | No | ||
| 2 | EGFR | 359 | 0.593 | 0.1072 | No | ||
| 3 | PPARGC1A | 3535 | 0.341 | 0.0289 | No | ||
| 4 | UBE2N | 4306 | 0.307 | 0.0320 | No | ||
| 5 | NDUFS4 | 4492 | 0.300 | 0.0550 | No | ||
| 6 | IL3 | 6077 | 0.239 | 0.0227 | No | ||
| 7 | PRKCG | 8378 | 0.163 | -0.0423 | No | ||
| 8 | SET | 8702 | 0.153 | -0.0387 | No | ||
| 9 | CARD14 | 9186 | 0.138 | -0.0420 | No | ||
| 10 | IL12A | 10297 | 0.104 | -0.0709 | No | ||
| 11 | LYN | 11511 | 0.070 | -0.1067 | No | ||
| 12 | TGFB1 | 11836 | 0.060 | -0.1122 | No | ||
| 13 | GTPBP4 | 11915 | 0.058 | -0.1092 | No | ||
| 14 | TDGF1 | 13472 | 0.015 | -0.1626 | No | ||
| 15 | IL5 | 14158 | -0.005 | -0.1862 | No | ||
| 16 | CD80 | 15229 | -0.035 | -0.2205 | No | ||
| 17 | AKTIP | 15532 | -0.043 | -0.2269 | No | ||
| 18 | IL22RA2 | 17052 | -0.085 | -0.2721 | No | ||
| 19 | CD81 | 17119 | -0.086 | -0.2659 | No | ||
| 20 | BRCA1 | 17535 | -0.098 | -0.2709 | No | ||
| 21 | CCND1 | 18562 | -0.127 | -0.2945 | No | ||
| 22 | PPP2R4 | 18977 | -0.141 | -0.2953 | No | ||
| 23 | ATG7 | 19412 | -0.154 | -0.2954 | No | ||
| 24 | RNF139 | 19796 | -0.165 | -0.2926 | No | ||
| 25 | IL31RA | 19907 | -0.169 | -0.2799 | No | ||
| 26 | ITLN1 | 21324 | -0.214 | -0.3087 | Yes | ||
| 27 | BCL10 | 21735 | -0.227 | -0.3008 | Yes | ||
| 28 | ITGB2 | 21745 | -0.228 | -0.2786 | Yes | ||
| 29 | UBE2D1 | 22066 | -0.239 | -0.2664 | Yes | ||
| 30 | IGFBP3 | 22264 | -0.246 | -0.2492 | Yes | ||
| 31 | BMP4 | 22546 | -0.255 | -0.2340 | Yes | ||
| 32 | NF2 | 24004 | -0.310 | -0.2548 | Yes | ||
| 33 | TSC1 | 24266 | -0.320 | -0.2325 | Yes | ||
| 34 | NLRP12 | 24360 | -0.325 | -0.2038 | Yes | ||
| 35 | STUB1 | 24893 | -0.346 | -0.1885 | Yes | ||
| 36 | CCND2 | 25943 | -0.402 | -0.1859 | Yes | ||
| 37 | TNK2 | 26170 | -0.416 | -0.1529 | Yes | ||
| 38 | IL29 | 26202 | -0.418 | -0.1128 | Yes | ||
| 39 | HCLS1 | 26232 | -0.419 | -0.0725 | Yes | ||
| 40 | CLCF1 | 26855 | -0.461 | -0.0491 | Yes | ||
| 41 | CCND3 | 27316 | -0.500 | -0.0161 | Yes | ||
| 42 | SOCS1 | 27851 | -0.561 | 0.0203 | Yes |