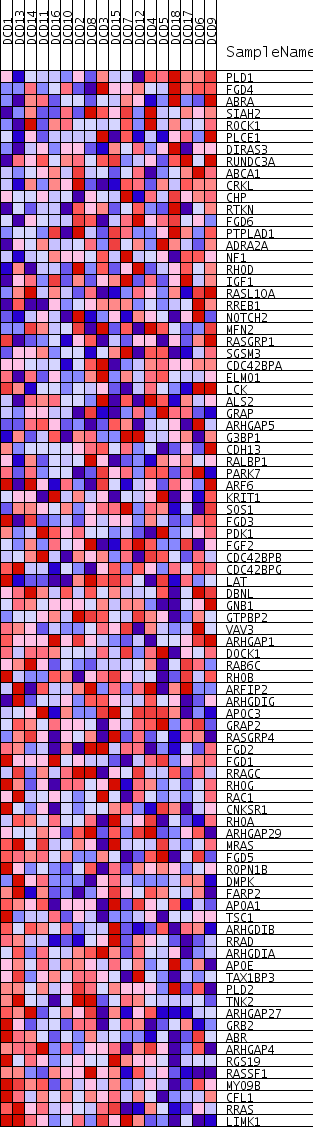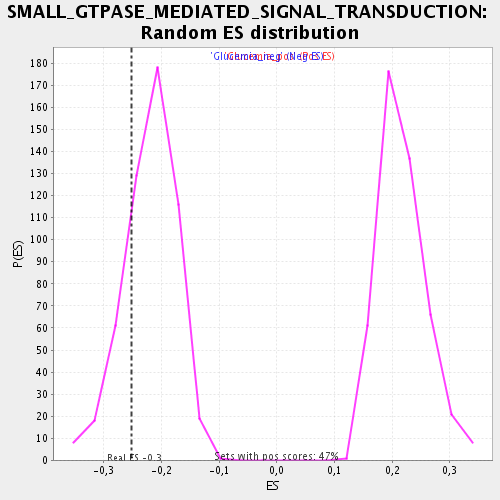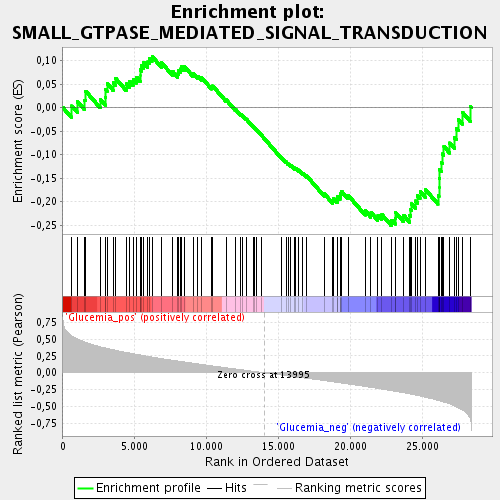
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | DCD_collapsed_to_symbols.DCD.cls#Glucemia |
| Phenotype | DCD.cls#Glucemia |
| Upregulated in class | Glucemia_neg |
| GeneSet | SMALL_GTPASE_MEDIATED_SIGNAL_TRANSDUCTION |
| Enrichment Score (ES) | -0.2515543 |
| Normalized Enrichment Score (NES) | -1.1495434 |
| Nominal p-value | 0.21132076 |
| FDR q-value | 0.7603031 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PLD1 | 615 | 0.545 | 0.0036 | No | ||
| 2 | FGD4 | 1004 | 0.504 | 0.0133 | No | ||
| 3 | ABRA | 1521 | 0.457 | 0.0163 | No | ||
| 4 | SIAH2 | 1576 | 0.452 | 0.0354 | No | ||
| 5 | ROCK1 | 2597 | 0.387 | 0.0173 | No | ||
| 6 | PLCE1 | 2939 | 0.370 | 0.0224 | No | ||
| 7 | DIRAS3 | 2977 | 0.368 | 0.0382 | No | ||
| 8 | RUNDC3A | 3072 | 0.363 | 0.0517 | No | ||
| 9 | ABCA1 | 3491 | 0.343 | 0.0529 | No | ||
| 10 | CRKL | 3671 | 0.334 | 0.0620 | No | ||
| 11 | CHP | 4397 | 0.304 | 0.0506 | No | ||
| 12 | RTKN | 4626 | 0.295 | 0.0562 | No | ||
| 13 | FGD6 | 4879 | 0.284 | 0.0605 | No | ||
| 14 | PTPLAD1 | 5112 | 0.276 | 0.0651 | No | ||
| 15 | ADRA2A | 5368 | 0.265 | 0.0684 | No | ||
| 16 | NF1 | 5371 | 0.265 | 0.0806 | No | ||
| 17 | RHOD | 5454 | 0.262 | 0.0899 | No | ||
| 18 | IGF1 | 5626 | 0.256 | 0.0957 | No | ||
| 19 | RASL10A | 5908 | 0.245 | 0.0972 | No | ||
| 20 | RREB1 | 6003 | 0.241 | 0.1051 | No | ||
| 21 | NOTCH2 | 6197 | 0.234 | 0.1091 | No | ||
| 22 | MFN2 | 6853 | 0.212 | 0.0958 | No | ||
| 23 | RASGRP1 | 7638 | 0.186 | 0.0768 | No | ||
| 24 | SGSM3 | 8002 | 0.174 | 0.0721 | No | ||
| 25 | CDC42BPA | 8040 | 0.173 | 0.0788 | No | ||
| 26 | ELMO1 | 8165 | 0.169 | 0.0823 | No | ||
| 27 | LCK | 8257 | 0.167 | 0.0868 | No | ||
| 28 | ALS2 | 8464 | 0.160 | 0.0870 | No | ||
| 29 | GRAP | 9047 | 0.142 | 0.0731 | No | ||
| 30 | ARHGAP5 | 9386 | 0.132 | 0.0673 | No | ||
| 31 | G3BP1 | 9646 | 0.124 | 0.0639 | No | ||
| 32 | CDH13 | 10336 | 0.103 | 0.0444 | No | ||
| 33 | RALBP1 | 10419 | 0.101 | 0.0461 | No | ||
| 34 | PARK7 | 11355 | 0.074 | 0.0166 | No | ||
| 35 | ARF6 | 11989 | 0.056 | -0.0031 | No | ||
| 36 | KRIT1 | 12366 | 0.045 | -0.0143 | No | ||
| 37 | SOS1 | 12486 | 0.042 | -0.0165 | No | ||
| 38 | FGD3 | 12747 | 0.035 | -0.0241 | No | ||
| 39 | PDK1 | 13228 | 0.022 | -0.0400 | No | ||
| 40 | FGF2 | 13339 | 0.019 | -0.0430 | No | ||
| 41 | CDC42BPB | 13431 | 0.016 | -0.0455 | No | ||
| 42 | CDC42BPG | 13834 | 0.004 | -0.0595 | No | ||
| 43 | LAT | 15195 | -0.034 | -0.1059 | No | ||
| 44 | DBNL | 15523 | -0.043 | -0.1155 | No | ||
| 45 | GNB1 | 15708 | -0.048 | -0.1197 | No | ||
| 46 | GTPBP2 | 15863 | -0.053 | -0.1227 | No | ||
| 47 | VAV3 | 16095 | -0.059 | -0.1282 | No | ||
| 48 | ARHGAP1 | 16191 | -0.062 | -0.1287 | No | ||
| 49 | DOCK1 | 16390 | -0.067 | -0.1325 | No | ||
| 50 | RAB6C | 16693 | -0.075 | -0.1397 | No | ||
| 51 | RHOB | 16945 | -0.082 | -0.1448 | No | ||
| 52 | ARFIP2 | 18188 | -0.116 | -0.1832 | No | ||
| 53 | ARHGDIG | 18763 | -0.133 | -0.1973 | No | ||
| 54 | APOC3 | 18845 | -0.136 | -0.1938 | No | ||
| 55 | GRAP2 | 19085 | -0.144 | -0.1956 | No | ||
| 56 | RASGRP4 | 19103 | -0.145 | -0.1895 | No | ||
| 57 | FGD2 | 19277 | -0.150 | -0.1886 | No | ||
| 58 | FGD1 | 19328 | -0.152 | -0.1834 | No | ||
| 59 | RRAGC | 19380 | -0.153 | -0.1780 | No | ||
| 60 | RHOG | 19836 | -0.167 | -0.1864 | No | ||
| 61 | RAC1 | 21030 | -0.204 | -0.2190 | No | ||
| 62 | CNKSR1 | 21430 | -0.218 | -0.2230 | No | ||
| 63 | RHOA | 21915 | -0.234 | -0.2292 | No | ||
| 64 | ARHGAP29 | 22190 | -0.243 | -0.2276 | No | ||
| 65 | MRAS | 22870 | -0.266 | -0.2392 | Yes | ||
| 66 | FGD5 | 23111 | -0.275 | -0.2349 | Yes | ||
| 67 | ROPN1B | 23146 | -0.277 | -0.2233 | Yes | ||
| 68 | DMPK | 23703 | -0.298 | -0.2291 | Yes | ||
| 69 | FARP2 | 24119 | -0.314 | -0.2291 | Yes | ||
| 70 | APOA1 | 24179 | -0.317 | -0.2165 | Yes | ||
| 71 | TSC1 | 24266 | -0.320 | -0.2047 | Yes | ||
| 72 | ARHGDIB | 24527 | -0.332 | -0.1985 | Yes | ||
| 73 | RRAD | 24660 | -0.337 | -0.1875 | Yes | ||
| 74 | ARHGDIA | 24844 | -0.344 | -0.1780 | Yes | ||
| 75 | APOE | 25235 | -0.364 | -0.1748 | Yes | ||
| 76 | TAX1BP3 | 26093 | -0.412 | -0.1860 | Yes | ||
| 77 | PLD2 | 26165 | -0.416 | -0.1692 | Yes | ||
| 78 | TNK2 | 26170 | -0.416 | -0.1500 | Yes | ||
| 79 | ARHGAP27 | 26208 | -0.418 | -0.1319 | Yes | ||
| 80 | GRB2 | 26339 | -0.426 | -0.1167 | Yes | ||
| 81 | ABR | 26384 | -0.428 | -0.0984 | Yes | ||
| 82 | ARHGAP4 | 26505 | -0.436 | -0.0824 | Yes | ||
| 83 | RGS19 | 26910 | -0.464 | -0.0751 | Yes | ||
| 84 | RASSF1 | 27247 | -0.494 | -0.0640 | Yes | ||
| 85 | MYO9B | 27363 | -0.506 | -0.0446 | Yes | ||
| 86 | CFL1 | 27505 | -0.522 | -0.0254 | Yes | ||
| 87 | RRAS | 27778 | -0.552 | -0.0094 | Yes | ||
| 88 | LIMK1 | 28357 | -0.696 | 0.0025 | Yes |